You are browsing environment: FUNGIDB
CAZyme Information: RAK81208.1
You are here: Home > Sequence: RAK81208.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
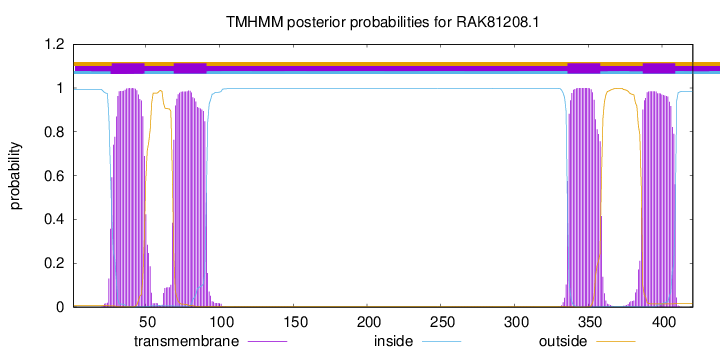
TMHMM annotations
Basic Information help
| Species | Aspergillus fijiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis | |||||||||||
| CAZyme ID | RAK81208.1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404513 | Glyco_trans_2_3 | 7.10e-12 | 191 | 361 | 1 | 167 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 224136 | BcsA | 3.02e-05 | 190 | 360 | 139 | 314 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.18e-107 | 19 | 420 | 14 | 451 | |
| 1.46e-85 | 17 | 420 | 27 | 444 | |
| 2.81e-84 | 17 | 420 | 27 | 427 | |
| 8.95e-61 | 17 | 336 | 5 | 331 | |
| 1.19e-54 | 93 | 336 | 75 | 328 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.98e-20 | 38 | 333 | 21 | 327 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis elegans OX=6239 GN=bre-3 PE=2 SV=2 |
|
| 4.41e-19 | 137 | 333 | 130 | 327 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis briggsae OX=6238 GN=bre-3 PE=3 SV=1 |
|
| 2.63e-18 | 170 | 333 | 161 | 327 | Beta-1,4-mannosyltransferase egh OS=Drosophila melanogaster OX=7227 GN=egh PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
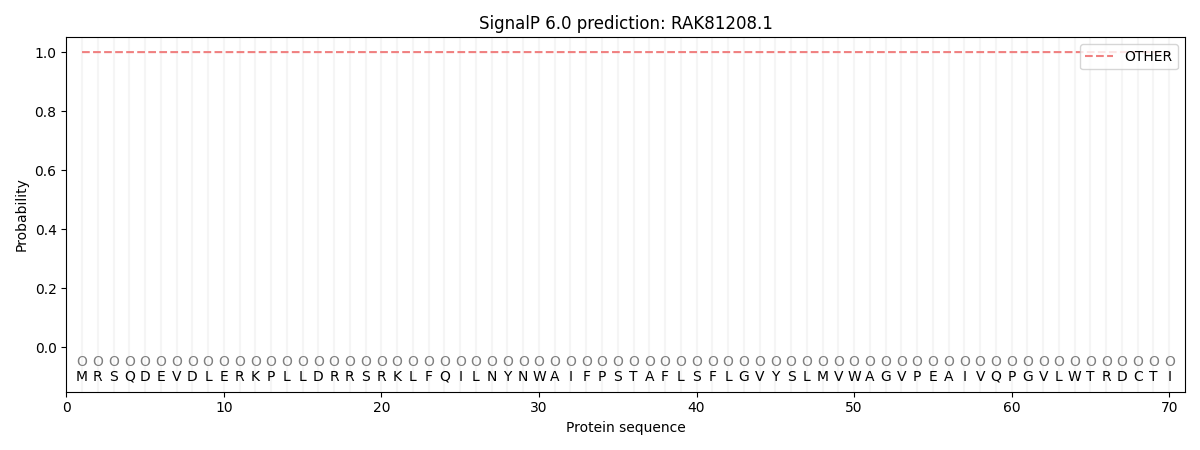
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000014 | 0.000011 |