You are browsing environment: FUNGIDB
CAZyme Information: RAK78915.1
Basic Information
help
| Species |
Aspergillus fijiensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis
|
| CAZyme ID |
RAK78915.1
|
| CAZy Family |
GH47 |
| CAZyme Description |
six-hairpin glycosidase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 683 |
KZ824635|CGC5 |
75883.81 |
4.7725 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AfijiensisCBS313.89 |
12336 |
1448319 |
318 |
12018
|
|
| Gene Location |
No EC number prediction in RAK78915.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH142 |
34 |
515 |
7.7e-156 |
0.9624217118997912 |
RAK78915.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
14 |
681 |
10 |
678 |
| 0.0 |
14 |
681 |
10 |
678 |
| 0.0 |
10 |
681 |
7 |
677 |
| 0.0 |
10 |
681 |
24 |
694 |
| 0.0 |
10 |
681 |
7 |
677 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.36e-69 |
38 |
516 |
653 |
1101 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 7.29e-66 |
38 |
516 |
653 |
1101 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
RAK78915.1 has no Swissprot hit.
This protein is predicted as SP
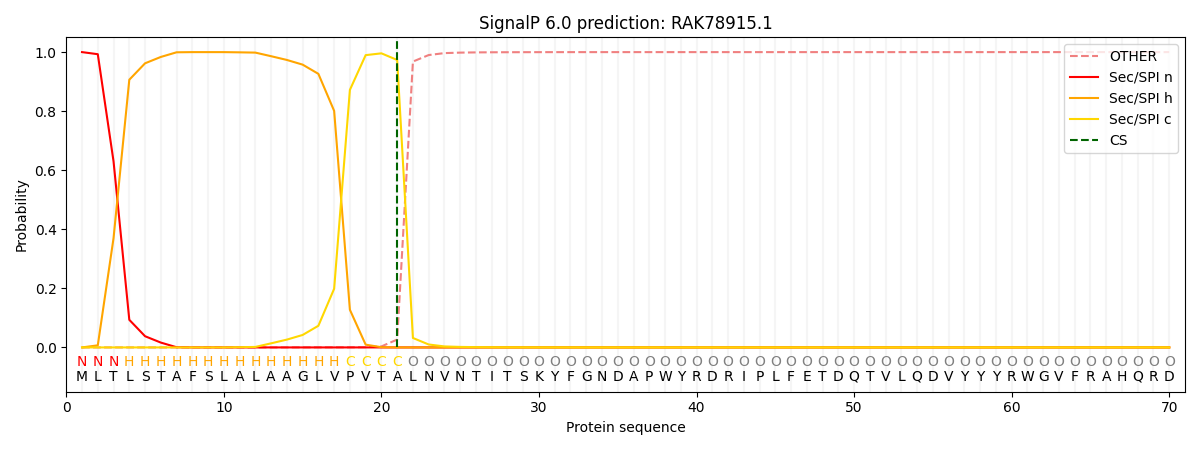
| Other |
SP_Sec_SPI |
CS Position |
| 0.000343 |
0.999619 |
CS pos: 21-22. Pr: 0.9734 |
There is no transmembrane helices in RAK78915.1.

