You are browsing environment: FUNGIDB
CAZyme Information: RAK78477.1
You are here: Home > Sequence: RAK78477.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus fijiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis | |||||||||||
| CAZyme ID | RAK78477.1 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | putative beta-galactosidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 47 | 393 | 1.4e-83 | 0.9869706840390879 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 1.27e-82 | 46 | 393 | 1 | 314 | Glycosyl hydrolases family 35. |
| 402180 | BetaGal_dom2 | 4.19e-81 | 403 | 582 | 1 | 179 | Beta-galactosidase, domain 2. This is the second domain of the five-domain beta-galactosidase enzyme that altogether catalyzes the hydrolysis of beta(1-3) and beta(1-4) galactosyl bonds in oligosaccharides as well as the inverse reaction of enzymatic condensation and trans-glycosylation. This domain is made up of 16 antiparallel beta-strands and an alpha-helix at its C-terminus. The fold of this domain appears to be unique. In addition, the last seven strands of the domain form a subdomain with an immunoglobulin-like (I-type Ig) fold in which the first strand is divided between the two beta-sheets. In penicillin spp this strand is interrupted by a 12-residue insertion which forms an additional edge-strand to the second beta-sheet of the sub-domain. The remainder of the second domain forms a series of beta-hairpins at its N-terminus, four strands of which are contiguous with part of the Ig-like sub-domain, forming in total a seven-stranded antiparallel beta-sheet. This domain is associated with family Glyco_hydro_35, pfam01301, which is N-terminal to it, but itself has no metazoan members. |
| 198097 | BetaGal_dom2 | 1.51e-66 | 402 | 584 | 1 | 182 | Beta-galactosidase, domain 2. This is the second domain of the five-domain beta-galactosidase enzyme that altogether catalyses the hydrolysis of beta(1-3) and beta(1-4) galactosyl bonds in oligosaccharides as well as the inverse reaction of enzymatic condensation and trans-glycosylation. This domain is made up of 16 antiparallel beta-strands and an alpha-helix at its C terminus. The fold of this domain appears to be unique. In addition, the last seven strands of the domain form a subdomain with an immunoglobulin-like (I-type Ig) fold in which the first strand is divided between the two beta-sheets. In penicillin spp this strand is interrupted by a 12-residue insertion which forms an additional edge-strand to the second beta-sheet of the sub-domain. The remainder of the second domain forms a series of beta-hairpins at its N terminus, four strands of which are contiguous with part of the Ig-like sub-domain, forming in total a seven-stranded antiparallel beta-sheet. This domain is associated with family Glyco_hydro_35, which is N-terminal to it, but itself has no metazoan members. |
| 404274 | BetaGal_dom4_5 | 3.01e-35 | 866 | 983 | 1 | 111 | Beta-galactosidase jelly roll domain. This domain is found in beta galactosidase enzymes. It has a jelly roll fold. |
| 166698 | PLN03059 | 2.85e-26 | 36 | 379 | 26 | 323 | beta-galactosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1023 | 1 | 1015 | |
| 0.0 | 10 | 1023 | 10 | 1014 | |
| 0.0 | 10 | 1023 | 10 | 1014 | |
| 0.0 | 22 | 1023 | 1 | 1001 | |
| 0.0 | 1 | 1023 | 1 | 1036 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.25e-187 | 40 | 985 | 6 | 936 | Native structure of beta-galactosidase from Penicillium sp. [Penicillium sp.],1XC6_A Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose [Penicillium sp.] |
|
| 5.23e-169 | 40 | 984 | 46 | 971 | Structure Of Beta-galactosidase From Aspergillus Niger [Aspergillus niger CBS 513.88] |
|
| 1.45e-168 | 40 | 984 | 46 | 971 | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose [Aspergillus niger CBS 513.88],5IHR_A Structure Of E298q-beta-galactosidase From Aspergillus Niger In Complex With Allolactose [Aspergillus niger CBS 513.88],5JUV_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose [Aspergillus niger CBS 513.88],5MGC_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose [Aspergillus niger CBS 513.88],5MGD_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose [Aspergillus niger CBS 513.88] |
|
| 4.58e-163 | 40 | 1010 | 46 | 994 | Crystal structure of beta-galactosidase from Aspergillus oryzae in complex with galactose [Aspergillus oryzae] |
|
| 2.36e-162 | 40 | 987 | 26 | 970 | Chain A, Beta-galactosidase [Trichoderma reesei],3OGR_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGS_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGV_A Chain A, Beta-galactosidase [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 1023 | 1 | 1013 | Probable beta-galactosidase B OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=lacB PE=3 SV=2 |
|
| 0.0 | 1 | 1023 | 1 | 1015 | Probable beta-galactosidase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=lacB PE=3 SV=1 |
|
| 0.0 | 1 | 1023 | 1 | 1022 | Probable beta-galactosidase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=lacB PE=3 SV=2 |
|
| 0.0 | 15 | 1023 | 21 | 1009 | Probable beta-galactosidase B OS=Pyrenophora tritici-repentis (strain Pt-1C-BFP) OX=426418 GN=lacB PE=3 SV=1 |
|
| 0.0 | 19 | 1023 | 21 | 1022 | Probable beta-galactosidase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=lacB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
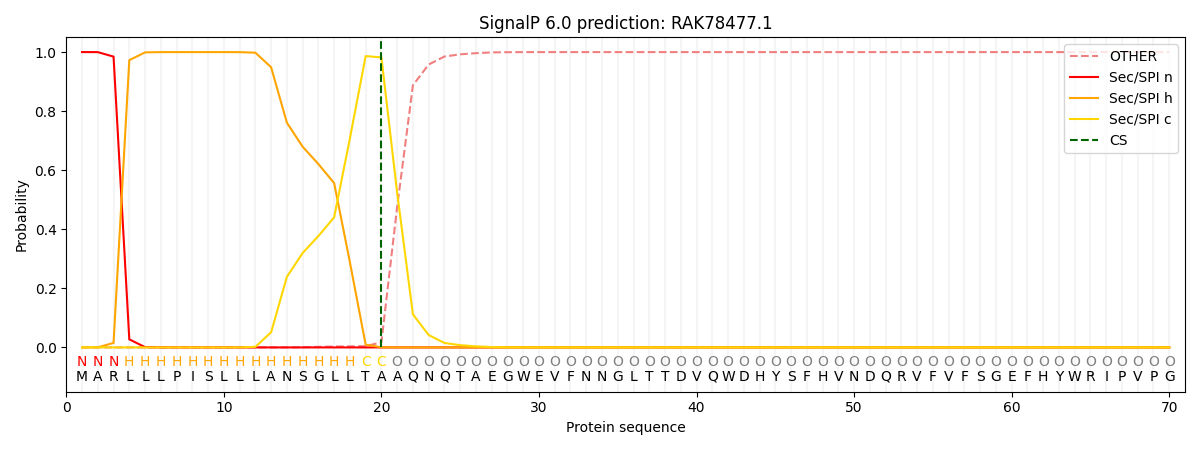
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000203 | 0.999819 | CS pos: 20-21. Pr: 0.9821 |
