You are browsing environment: FUNGIDB
CAZyme Information: RAK76512.1
You are here: Home > Sequence: RAK76512.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
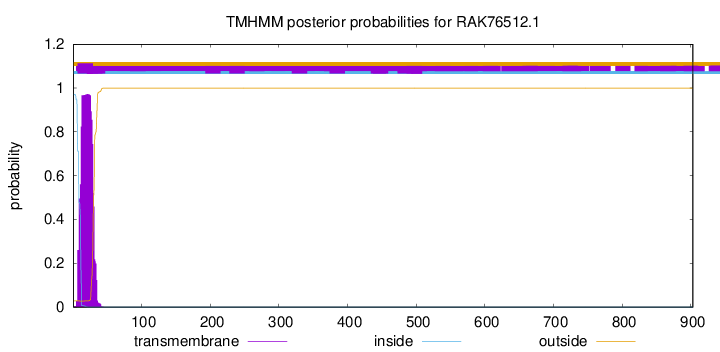
TMHMM annotations
Basic Information help
| Species | Aspergillus fijiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis | |||||||||||
| CAZyme ID | RAK76512.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | putative beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.165:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 57 | 733 | 2.3e-51 | 0.663563829787234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408168 | Ig_GlcNase | 3.47e-36 | 785 | 895 | 1 | 104 | Exo-beta-D-glucosaminidase Ig-fold domain. This domain can be found in 2 glycoside hydrolase subfamily of beta-glucosaminidases (EC:3.2.1.165) such as CsxA, from Amycolatopsis orientalis that has exo-beta-D-glucosaminidase (exo-chitosanase) activity. It has an immunoglobulin-like topology. |
| 225789 | LacZ | 7.15e-14 | 120 | 661 | 98 | 623 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 236673 | ebgA | 3.16e-10 | 107 | 455 | 112 | 452 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| 395572 | Glyco_hydro_2 | 3.80e-06 | 219 | 333 | 6 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| 236657 | PRK10150 | 2.03e-05 | 90 | 398 | 51 | 353 | beta-D-glucuronidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 29 | 902 | 22 | 879 | |
| 0.0 | 29 | 902 | 22 | 879 | |
| 0.0 | 29 | 902 | 22 | 879 | |
| 0.0 | 29 | 902 | 22 | 879 | |
| 0.0 | 29 | 902 | 22 | 879 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.15e-167 | 31 | 884 | 49 | 876 | Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZS_B Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2X05_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X05_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_A Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis],2X09_B Inhibition of the exo-beta-D-glucosaminidase CsxA by a glucosamine- configured castanospermine and an amino-australine analogue [Amycolatopsis orientalis] |
|
| 2.78e-166 | 31 | 884 | 49 | 876 | Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis],2VZO_B Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA [Amycolatopsis orientalis] |
|
| 5.50e-166 | 31 | 884 | 49 | 876 | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZT_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZV_A Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis],2VZV_B Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan [Amycolatopsis orientalis] |
|
| 1.64e-164 | 31 | 884 | 49 | 876 | Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis],2VZU_B Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine [Amycolatopsis orientalis] |
|
| 4.02e-19 | 106 | 667 | 82 | 652 | Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.72e-281 | 31 | 900 | 33 | 882 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
|
| 3.41e-262 | 31 | 900 | 30 | 890 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
|
| 4.12e-260 | 31 | 900 | 28 | 888 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
|
| 6.72e-174 | 27 | 900 | 45 | 901 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
|
| 2.62e-166 | 31 | 884 | 49 | 876 | Exo-beta-D-glucosaminidase OS=Amycolatopsis orientalis OX=31958 GN=csxA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.139717 | 0.860246 | CS pos: 25-26. Pr: 0.8288 |