You are browsing environment: FUNGIDB
CAZyme Information: RAK76363.1
You are here: Home > Sequence: RAK76363.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus fijiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis | |||||||||||
| CAZyme ID | RAK76363.1 | |||||||||||
| CAZy Family | GH142 | |||||||||||
| CAZyme Description | glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 391 | 627 | 7.6e-21 | 0.9196141479099679 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.30e-54 | 371 | 629 | 44 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 236766 | rne | 8.13e-07 | 27 | 213 | 852 | 1009 | ribonuclease E; Reviewed |
| 236766 | rne | 7.34e-05 | 58 | 326 | 841 | 1060 | ribonuclease E; Reviewed |
| 223021 | PHA03247 | 7.65e-05 | 94 | 213 | 2762 | 2881 | large tegument protein UL36; Provisional |
| 411345 | gliding_GltJ | 1.39e-04 | 92 | 200 | 415 | 521 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.30e-188 | 1 | 629 | 1 | 630 | |
| 5.30e-188 | 1 | 629 | 1 | 630 | |
| 2.13e-169 | 1 | 629 | 1 | 601 | |
| 2.13e-169 | 1 | 629 | 1 | 601 | |
| 2.13e-169 | 1 | 629 | 1 | 601 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.78e-170 | 1 | 629 | 1 | 601 | Probable beta-glucosidase btgE OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgE PE=3 SV=1 |
|
| 3.78e-170 | 1 | 629 | 1 | 601 | Probable beta-glucosidase btgE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgE PE=3 SV=1 |
|
| 5.94e-142 | 1 | 629 | 1 | 564 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
|
| 1.38e-135 | 1 | 629 | 1 | 558 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 6.84e-135 | 1 | 629 | 1 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
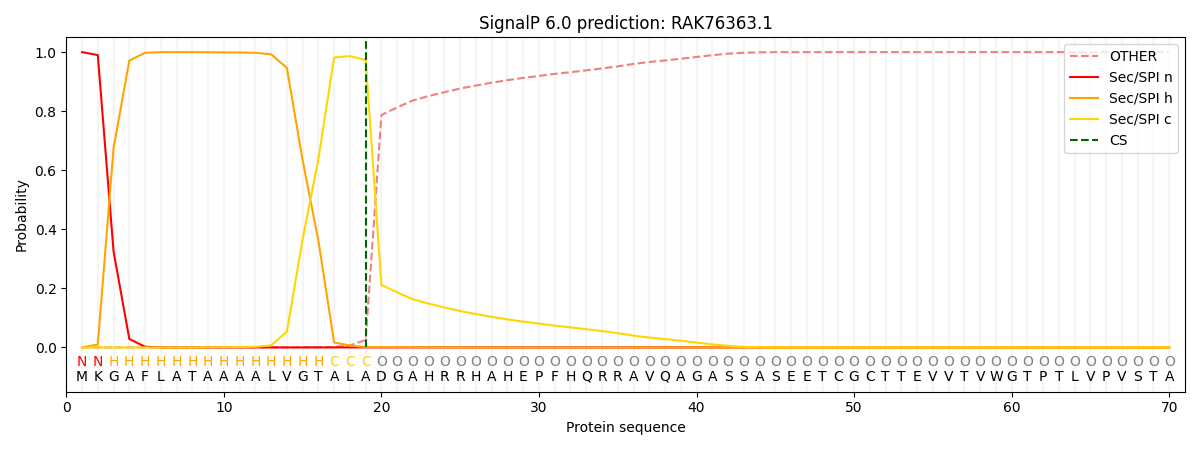
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000804 | 0.999152 | CS pos: 19-20. Pr: 0.9733 |
