You are browsing environment: FUNGIDB
CAZyme Information: RAK73495.1
You are here: Home > Sequence: RAK73495.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus fijiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fijiensis | |||||||||||
| CAZyme ID | RAK73495.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | cutinase-domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.74:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 45 | 221 | 1.8e-40 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 1.70e-53 | 45 | 222 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.70e-81 | 1 | 225 | 1 | 224 | |
| 2.42e-81 | 1 | 225 | 1 | 224 | |
| 2.42e-81 | 1 | 225 | 1 | 224 | |
| 2.42e-81 | 1 | 225 | 1 | 224 | |
| 5.02e-81 | 7 | 225 | 8 | 227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.10e-57 | 29 | 222 | 5 | 196 | Crystal structure of Aspergillus oryzae cutinase [Aspergillus oryzae] |
|
| 6.88e-56 | 39 | 222 | 5 | 187 | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon [Aspergillus oryzae] |
|
| 1.14e-54 | 35 | 223 | 12 | 199 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 1.65e-50 | 33 | 222 | 1 | 192 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
|
| 6.84e-50 | 35 | 222 | 3 | 193 | Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.29e-82 | 1 | 225 | 1 | 224 | Probable cutinase 1 OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=AFLA_039350 PE=3 SV=1 |
|
| 1.23e-81 | 1 | 225 | 1 | 224 | Probable cutinase 3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=AO090011000113 PE=3 SV=1 |
|
| 8.52e-81 | 1 | 225 | 1 | 227 | Probable cutinase 1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An14g02170 PE=3 SV=1 |
|
| 8.06e-78 | 3 | 222 | 2 | 215 | Probable cutinase 3 OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=ACLA_055320 PE=3 SV=1 |
|
| 7.00e-77 | 7 | 222 | 6 | 215 | Probable cutinase 3 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_030250 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
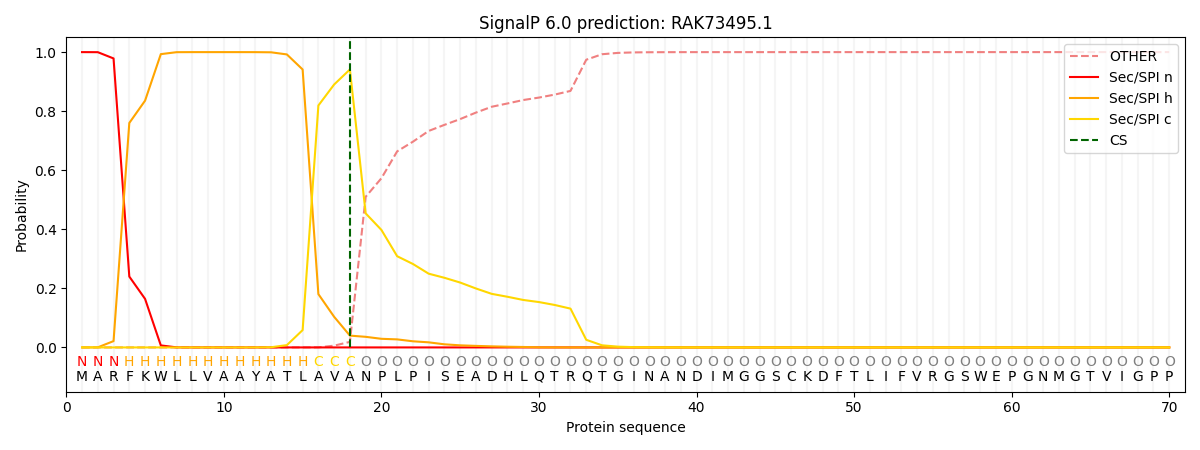
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000222 | 0.999732 | CS pos: 18-19. Pr: 0.9412 |
