You are browsing environment: FUNGIDB
CAZyme Information: QYA_78557T0-p1
You are here: Home > Sequence: QYA_78557T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
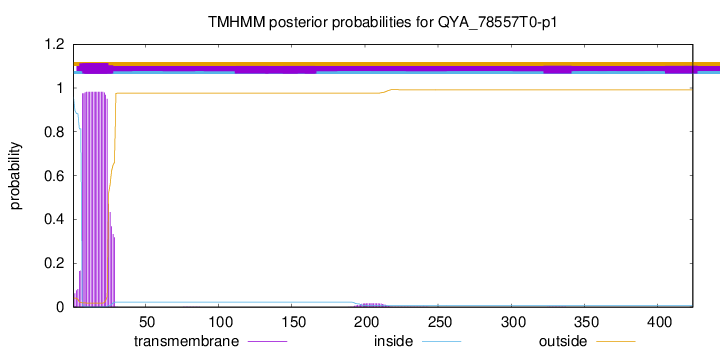
TMHMM annotations
Basic Information help
| Species | Mucor lusitanicus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor lusitanicus | |||||||||||
| CAZyme ID | QYA_78557T0-p1 | |||||||||||
| CAZy Family | PL38 | |||||||||||
| CAZyme Description | Glycosyltransferase family 49 protein [Source:UniProtKB/TrEMBL;Acc:A0A168NKE2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404735 | Glyco_transf_49 | 3.77e-16 | 99 | 369 | 3 | 312 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.57e-132 | 4 | 416 | 74 | 487 | |
| 5.61e-126 | 5 | 414 | 65 | 492 | |
| 9.86e-63 | 26 | 404 | 121 | 498 | |
| 3.36e-57 | 56 | 404 | 172 | 513 | |
| 4.02e-50 | 51 | 402 | 68 | 413 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
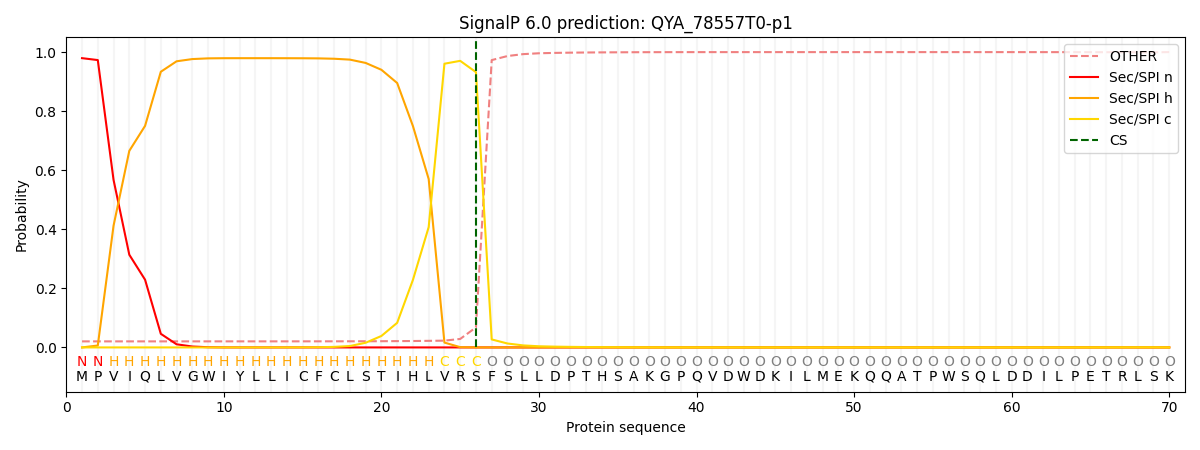
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.027312 | 0.972665 | CS pos: 26-27. Pr: 0.9322 |