You are browsing environment: FUNGIDB
CAZyme Information: QYA_77324T0-p1
Basic Information
help
| Species |
Mucor lusitanicus
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor lusitanicus
|
| CAZyme ID |
QYA_77324T0-p1
|
| CAZy Family |
GT57 |
| CAZyme Description |
Glycosyltransferase family 77 protein [Source:UniProtKB/TrEMBL;Acc:A0A162TWP8]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 346 |
|
39997.57 |
9.9360 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_MlusitanicusCBS277-49 |
11852 |
747725 |
0 |
11852
|
|
| Gene Location |
No EC number prediction in QYA_77324T0-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT77 |
131 |
335 |
1.3e-25 |
0.9675925925925926 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 367480
|
Nucleotid_trans |
1.33e-17 |
131 |
335 |
4 |
204 |
Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.29e-84 |
57 |
346 |
80 |
356 |
| 3.80e-78 |
42 |
346 |
77 |
368 |
| 2.57e-64 |
26 |
346 |
73 |
370 |
| 6.92e-64 |
100 |
346 |
87 |
333 |
| 3.59e-52 |
72 |
346 |
92 |
353 |
QYA_77324T0-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.35e-15 |
99 |
346 |
10 |
290 |
UDP-galactose:fucoside alpha-3-galactosyltransferase OS=Dictyostelium discoideum OX=44689 GN=agtA PE=1 SV=1 |
This protein is predicted as OTHER
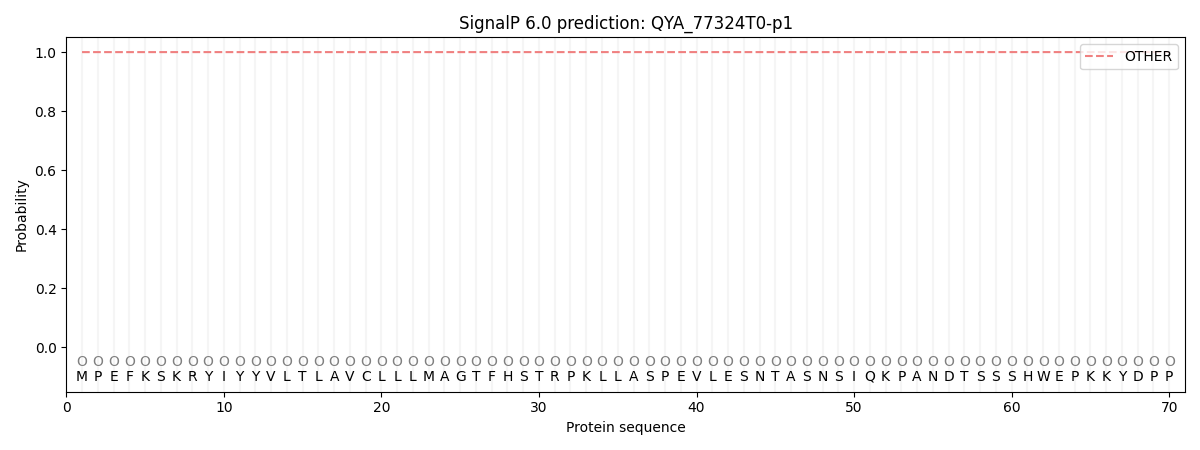
| Other |
SP_Sec_SPI |
CS Position |
| 0.999962 |
0.000069 |
|