You are browsing environment: FUNGIDB
CAZyme Information: QYA_181082T0-p1
You are here: Home > Sequence: QYA_181082T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mucor lusitanicus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor lusitanicus | |||||||||||
| CAZyme ID | QYA_181082T0-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | Glycoside hydrolase family 5 protein [Source:UniProtKB/TrEMBL;Acc:A0A162YLN3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 59 | 388 | 6.8e-133 | 0.9969135802469136 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 408348 | Glyco_hydro_5_C | 3.95e-20 | 408 | 494 | 1 | 86 | Glycoside hydrolase family 5 C-terminal domain. This is the C-terminal domain of endo-glycoceramidase II (EGC), a membrane-associated family 5 glycosidase pfam00150. The C-terminal domain assumes a beta-sandwich fold, which resembles that of many carbohydrate-binding modules. |
| 395098 | Cellulase | 5.10e-10 | 92 | 388 | 33 | 268 | Cellulase (glycosyl hydrolase family 5). |
| 396834 | Glyco_hydro_42 | 1.87e-04 | 87 | 148 | 14 | 85 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.07e-238 | 26 | 498 | 26 | 500 | |
| 1.13e-235 | 13 | 496 | 12 | 497 | |
| 1.13e-235 | 13 | 496 | 12 | 497 | |
| 1.36e-234 | 28 | 498 | 28 | 500 | |
| 3.36e-234 | 10 | 498 | 6 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.31e-26 | 35 | 461 | 36 | 446 | Chain A, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S],5CCU_B Chain B, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S] |
|
| 3.69e-25 | 35 | 461 | 36 | 446 | Chain A, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S],5J14_B Chain B, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S],5J7Z_A Chain A, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S],5J7Z_B Chain B, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S] |
|
| 2.29e-21 | 50 | 480 | 37 | 464 | Chain A, Endoglycoceramidase II [Rhodococcus sp.] |
|
| 4.11e-21 | 50 | 480 | 37 | 464 | Endo-glycoceramidase II from Rhodococcus sp. [Rhodococcus sp.],2OSW_B Endo-glycoceramidase II from Rhodococcus sp. [Rhodococcus sp.],2OYK_A Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex [Rhodococcus sp.],2OYK_B Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex [Rhodococcus sp.],2OYL_A Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex [Rhodococcus sp.],2OYL_B Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex [Rhodococcus sp.],2OYM_A Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex [Rhodococcus sp.],2OYM_B Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex [Rhodococcus sp.] |
|
| 5.22e-13 | 41 | 417 | 13 | 416 | Chain A, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-85 | 43 | 496 | 34 | 500 | Endoglycoceramidase OS=Cyanea nozakii OX=135523 PE=1 SV=1 |
|
| 5.41e-80 | 43 | 464 | 25 | 454 | Endoglycoceramidase OS=Hydra vulgaris OX=6087 PE=1 SV=1 |
|
| 5.43e-25 | 50 | 461 | 43 | 438 | Endoglycoceramidase I OS=Rhodococcus hoagii (strain 103S) OX=685727 GN=REQ_38260 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
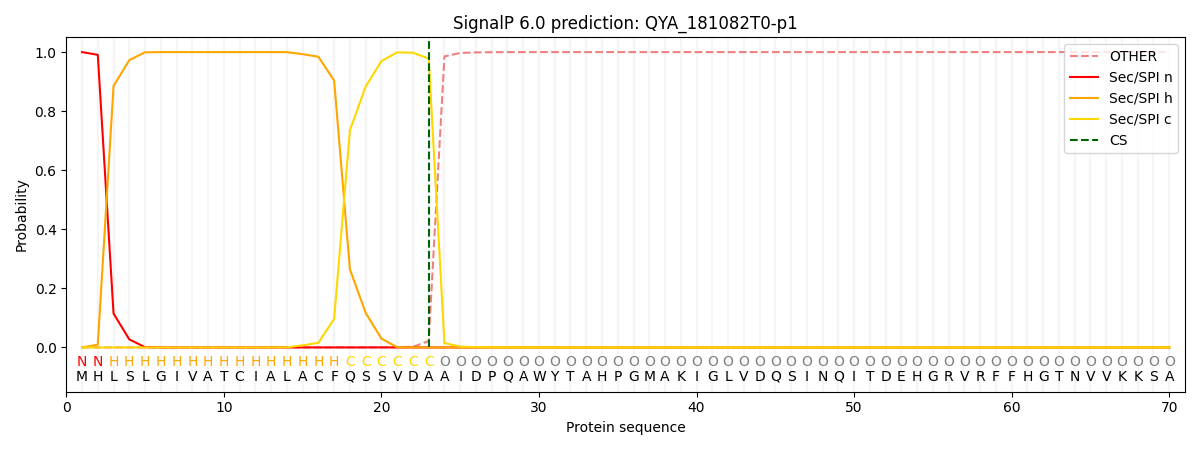
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000191 | 0.999799 | CS pos: 23-24. Pr: 0.9782 |
