You are browsing environment: FUNGIDB
CAZyme Information: QYA_143537T0-p1
You are here: Home > Sequence: QYA_143537T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
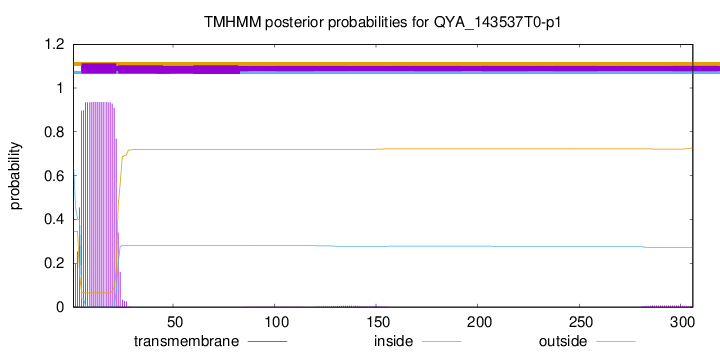
TMHMM annotations
Basic Information help
| Species | Mucor lusitanicus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor lusitanicus | |||||||||||
| CAZyme ID | QYA_143537T0-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Glyco_hydro_35 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A162TEA4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 55 | 259 | 2.2e-56 | 0.5928338762214984 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 5.05e-41 | 54 | 254 | 1 | 182 | Glycosyl hydrolases family 35. |
| 224786 | GanA | 3.17e-20 | 51 | 216 | 4 | 160 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 166698 | PLN03059 | 6.40e-14 | 50 | 216 | 32 | 186 | beta-galactosidase; Provisional |
| 396834 | Glyco_hydro_42 | 1.89e-05 | 76 | 217 | 8 | 138 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| 395098 | Cellulase | 0.005 | 55 | 116 | 1 | 62 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.25e-158 | 2 | 292 | 3 | 290 | |
| 3.57e-48 | 50 | 267 | 41 | 243 | |
| 1.70e-47 | 33 | 267 | 27 | 246 | |
| 1.74e-47 | 50 | 267 | 30 | 232 | |
| 2.39e-47 | 50 | 269 | 38 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.44e-28 | 50 | 291 | 48 | 260 | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose [Aspergillus niger CBS 513.88],5IHR_A Structure Of E298q-beta-galactosidase From Aspergillus Niger In Complex With Allolactose [Aspergillus niger CBS 513.88],5JUV_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose [Aspergillus niger CBS 513.88],5MGC_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose [Aspergillus niger CBS 513.88],5MGD_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose [Aspergillus niger CBS 513.88] |
|
| 2.44e-28 | 50 | 291 | 48 | 260 | Structure Of Beta-galactosidase From Aspergillus Niger [Aspergillus niger CBS 513.88] |
|
| 2.47e-28 | 48 | 257 | 18 | 209 | Chain A, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
|
| 1.25e-27 | 40 | 254 | 27 | 223 | Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
|
| 1.63e-27 | 40 | 254 | 3 | 199 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.29e-38 | 48 | 291 | 37 | 262 | Probable beta-galactosidase B OS=Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) OX=665079 GN=lacB PE=3 SV=1 |
|
| 1.75e-36 | 48 | 291 | 37 | 262 | Probable beta-galactosidase B OS=Botryotinia fuckeliana (strain B05.10) OX=332648 GN=lacB PE=3 SV=1 |
|
| 4.82e-33 | 46 | 291 | 26 | 247 | Probable beta-galactosidase C OS=Sclerotinia sclerotiorum (strain ATCC 18683 / 1980 / Ss-1) OX=665079 GN=lacC PE=3 SV=1 |
|
| 4.87e-33 | 48 | 291 | 40 | 264 | Probable beta-galactosidase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=lacB PE=3 SV=2 |
|
| 6.60e-33 | 48 | 293 | 40 | 267 | Probable beta-galactosidase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=lacB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
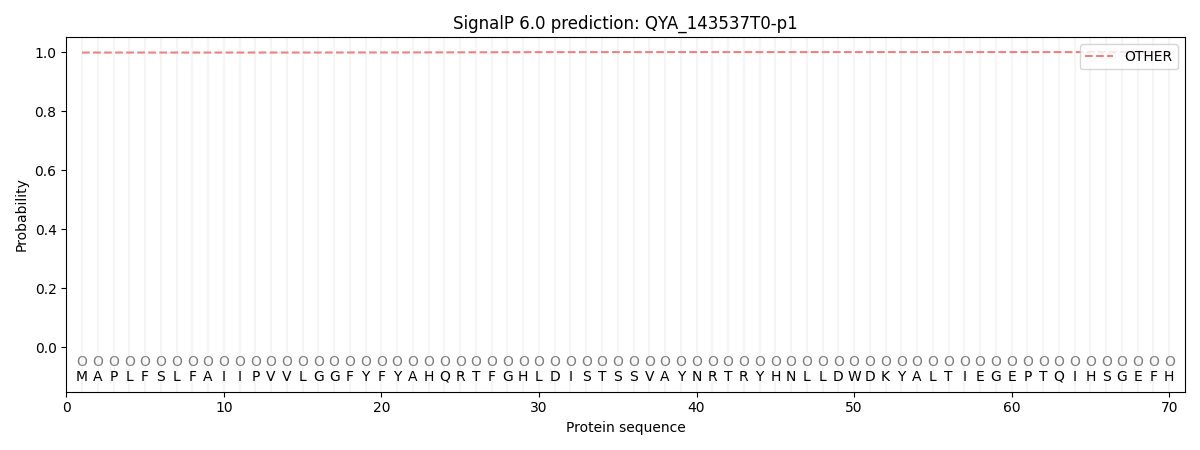
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998562 | 0.001482 |