You are browsing environment: FUNGIDB
CAZyme Information: QYA_127973T0-p1
You are here: Home > Sequence: QYA_127973T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mucor lusitanicus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Mucoraceae; Mucor; Mucor lusitanicus | |||||||||||
| CAZyme ID | QYA_127973T0-p1 | |||||||||||
| CAZy Family | GH125 | |||||||||||
| CAZyme Description | Mannosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A168JWW2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 1 | 403 | 2.4e-102 | 0.9974293059125964 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 9.18e-112 | 1 | 495 | 41 | 541 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 3.11e-80 | 1 | 403 | 2 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.27e-152 | 57 | 496 | 1 | 434 | |
| 6.91e-114 | 1 | 495 | 41 | 541 | |
| 6.91e-114 | 1 | 495 | 41 | 541 | |
| 6.91e-114 | 1 | 495 | 41 | 541 | |
| 6.91e-114 | 1 | 495 | 41 | 541 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-114 | 1 | 495 | 41 | 541 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
|
| 4.86e-109 | 1 | 486 | 55 | 522 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
|
| 1.56e-107 | 9 | 486 | 71 | 532 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
|
| 1.74e-106 | 9 | 486 | 60 | 521 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
|
| 2.15e-105 | 1 | 495 | 18 | 508 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.988997 | 0.010988 |
TMHMM Annotations download full data without filtering help
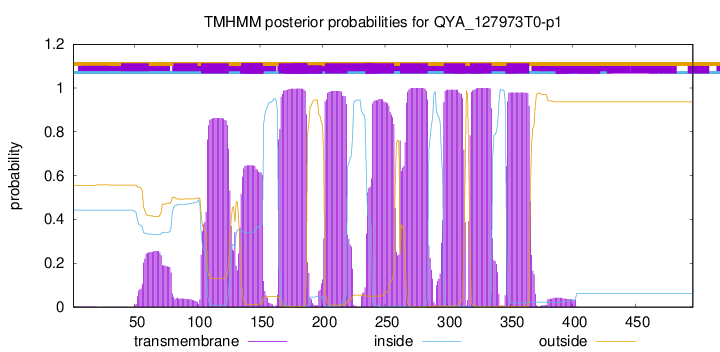
| Start | End |
|---|---|
| 103 | 125 |
| 135 | 152 |
| 165 | 187 |
| 202 | 219 |
| 235 | 257 |
| 262 | 284 |
| 297 | 314 |
| 318 | 335 |
| 347 | 365 |
