You are browsing environment: FUNGIDB
CAZyme Information: QSL66615.1
You are here: Home > Sequence: QSL66615.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis wakefieldiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis wakefieldiae | |||||||||||
| CAZyme ID | QSL66615.1 | |||||||||||
| CAZy Family | GT50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 99908; End:105775 Strand: + | |||||||||||
Full Sequence Download help
| MFAFDGIYKE LILFFILLQL RFFVALTSKT FFQPDEYWQS LEPAHQFVYG YGYLTWEWRE | 60 |
| GVRDFVYPFI FSIVYWLLKL TKYDSSYLII IFPKLLQAIF AAISDFYTIK FSLKLHGKRV | 120 |
| SFWTSIIVIT SPFNIFFGYR TFSNTLETVF IICALYYWPL NFKDFRKKEL RFSLIFLSIA | 180 |
| CILRPTNILI WLFLLGCLFF CGNFKFIFDV FIIGFISFLF LFLLNFYYYS RWTLPFVKFL | 240 |
| KFNLIDNSSV YYGSSPWHYY LFQGLPILLF LYLPFGLHGA YIHRKTIYFR LIVFILFCYS | 300 |
| MIQHKEIRFI YFLSPLLYIF SAKSISLVPM FIVKKMVSLI LIINVIVIWY FNQVHQRGVI | 360 |
| DVMGYIRNNL EVSGVVFLMP CHSTPWQSYI HRPDIQMRFL TCEPPINLHQ NNISIKYRDE | 420 |
| ADRFYDDPYL FLQKYFAYES LNSTLQDGFF WPSHFVFFEN LLAIVDSHIS ILGYKECARF | 480 |
| FNSHFIDDWR RKGYNFIVQP SYQLNSLRKY TMSTIQADRK VLPTNIILLI MGGIPSQSIT | 540 |
| YEKDEQTATF RFFESIPSGV DIKLVIKFRG EINNKMCGFY RSSYQDNKTK EIKWLACTQM | 600 |
| EATDCRRAFP CWDEPALKAT FDVEITADVR YTILSNMDIF RESILDDKRT VLFSRTPLMS | 660 |
| TYLLAWIIGE LDYIEMFTSG INMPKIPVRV YSPLPNLSQD GKFSLELAVK TLDFFSKTFG | 720 |
| IPYCLPKLDM VAIPDFSAGA MENWGIVTYR VVDLLFNEKT CSAATKQRVA EVVQHELAHQ | 780 |
| WFGDLVTMDF WDGLWLNEGF ATWMSWFSCN TFYPEWKVWQ TYITGSLQNA LRFDGFRSSH | 840 |
| PIQVLVKSAE EINQIFDAIS YSKGSSVIRM LSKYLGEDVF LAGIRRYLKH HSYGNATTDD | 900 |
| LWKALSDESG KDILKFMKCW TENVGYPVLT VSENDDSIIV RQNRFLASGD VKPEDDTTIY | 960 |
| WVPLMLKTMS IDGKITTDSY LILDKRESCI PMSNIKKGFY KINTGHSGIY RTLYPSKHLQ | 1020 |
| RLDRVGLVAD TGSLAESGYC RTSDLLTLIL GWHNEDNFVV WNETITRLEN LEAAFVFESE | 1080 |
| QIKHALSLYK RTLVSDKVRK LGWDFDANIS HIERQFKTLM FGTAGKAGDE EIISVAKDML | 1140 |
| YRYADGDMSA IHPDLRANVF GICIQYGSEK EWDIIYSISK NTTSSDEKNT ALKQLGLTRV | 1200 |
| PHLISRTLKL ILSSEVKLQD IYMPLIGLKT HSEGIKALWK FCNENWDGID NMIPSTVGSL | 1260 |
| KSTVVQLMVS GFTSMSSVHE IESFFKDKKI QAFDKALSQS LDAIRTKASW VSRDYNDVET | 1320 |
| WLKNNGYLNT I | 1331 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 14 | 391 | 8.3e-94 | 0.9948586118251928 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 341064 | M1_APN-Q_like | 0.0 | 533 | 922 | 54 | 442 | Peptidase M1 aminopeptidase N catalytic domain family which includes aminopeptidase N (APN), aminopeptidase Q (APQ), tricorn interacting factor F3, and endoplasmic reticulum aminopeptidase 1 (ERAP1). This M1 peptidase family includes eukaryotic and bacterial members: the catalytic domains of aminopeptidase N (APN), aminopeptidase Q (APQ, laeverin), endoplasmic reticulum aminopeptidase 1 (ERAP1) as well as tricorn interacting factor F3. Aminopeptidase N (APN; CD13; alanyl aminopeptidase; EC 3.4.11.2), a type II integral membrane protease, preferentially cleaves neutral amino acids from the N-terminus of oligopeptides and is present in a variety of human tissues and cell types (leukocyte, fibroblast, endothelial and epithelial cells). APN expression is dysregulated in inflammatory diseases such as chronic pain, rheumatoid arthritis, multiple sclerosis, systemic sclerosis, systemic lupus erythematosus, polymyositis/dermatomyosytis and pulmonary sarcoidosis, and is enhanced in tumor cells such as melanoma, renal, prostate, pancreas, colon, gastric and thyroid cancers. It is considered a marker of differentiation since it is predominantly expressed on stem cells and on cells of the granulocytic and monocytic lineages at distinct stages of differentiation. Thus, APN inhibition may lead to the development of anti-cancer and anti-inflammatory drugs. ERAP1, also known as endoplasmic reticulum aminopeptidase associated with antigen processing (ERAAP), adipocyte derived leucine aminopeptidase (A-LAP), or aminopeptidase regulating tumor necrosis factor receptor I (THFRI) shedding (ARTS-1), associates with the closely related ER aminopeptidase ERAP2, for the final trimming of peptides within the ER for presentation by MHC class I molecules. ERAP1 is associated with ankylosing spondylitis (AS), an inflammatory arthritis that predominantly affects the spine. ERAP1 also aids in the shedding of membrane-bound cytokine receptors. The tricorn interacting factor F3, together with factors F1 and F2, degrades the tricorn protease products, producing free amino acids, thus completing the proteasomal degradation pathway. F3 is homologous to F2, but not F1, and shows a strong preference for glutamate in the P1' position. APQ, also known as laeverin, is specifically expressed in human embryo-derived extravillous trophoblasts (EVTs) that invade the uterus during early placentation. It cleaves the N-terminal amino acid of various peptides such as angiotensin III, endokinin C, and kisspeptin-10, all expressed in the placenta in large quantities. APN is a receptor for coronaviruses, although the virus receptor interaction site seems to be distinct from the enzymatic site and aminopeptidase activity is not necessary for viral infection. APNs are also putative Cry toxin receptors. Cry1 proteins are pore-forming toxins that bind to the midgut epithelial cell membrane of susceptible insect larvae, causing extensive damage. Several different toxins, including Cry1Aa, Cry1Ab, Cry1Ac, Cry1Ba, Cry1Ca and Cry1Fa, have been shown to bind to APNs; however, a direct role of APN in cytotoxicity has been yet to be firmly established. |
| 223385 | PepN | 3.93e-170 | 540 | 1317 | 85 | 859 | Aminopeptidase N [Amino acid transport and metabolism]. |
| 396150 | Peptidase_M1 | 7.84e-101 | 703 | 920 | 1 | 220 | Peptidase family M1 domain. Members of this family are aminopeptidases. The members differ widely in specificity, hydrolysing acidic, basic or neutral N-terminal residues. This family includes leukotriene-A4 hydrolase, this enzyme also has an aminopeptidase activity. |
| 341065 | M1_APN | 1.94e-94 | 598 | 920 | 120 | 437 | Peptidase M1 family including aminopeptidase N catalytic domain. This model represents the catalytic domain of bacterial and eukaryotic aminopeptidase N (APN; CD13; alanyl aminopeptidase; EC 3.4.11.2), a type II integral membrane protease belonging to the M1 gluzincin family. APN preferentially cleaves neutral amino acids from the N-terminus of oligopeptides and, in higher eukaryotes, is present in a variety of human tissues and cell types (leukocyte, fibroblast, endothelial and epithelial cells). APN expression is dysregulated in inflammatory diseases such as chronic pain, rheumatoid arthritis, multiple sclerosis, systemic sclerosis, systemic lupus erythematosus, polymyositis/dermatomyosytis and pulmonary sarcoidosis, and is enhanced in tumor cells such as melanoma, renal, prostate, pancreas, colon, gastric and thyroid cancers. It is predominantly expressed on stem cells and on cells of the granulocytic and monocytic lineages at distinct stages of differentiation, thus considered a marker of differentiation. Thus, APN inhibition may lead to the development of anti-cancer and anti-inflammatory drugs. APNs are also present in many pathogenic bacteria and represent potential drug targets. Some APNs have been used commercially, such as one from Lactococcus lactis used in the food industry. APN also serves as a receptor for coronaviruses, although the virus receptor interaction site seems to be distinct from the enzymatic site and aminopeptidase activity is not necessary for viral infection. APNs have also been extensively studied as putative Cry toxin receptors. Cry1 proteins are pore-forming toxins that bind to the midgut epithelial cell membrane of susceptible insect larvae, causing extensive damage. Several different toxins, including Cry1Aa, Cry1Ab, Cry1Ac, Cry1Ba, Cry1Ca and Cry1Fa, have been shown to bind to APNs; however, a direct role of APN in cytotoxicity has been yet to be firmly established. |
| 341058 | M1 | 7.85e-90 | 549 | 907 | 64 | 413 | Peptidase M1 family includes the catalytic domains of aminopeptidase N and leukotriene A4 hydrolase. The model represents the catalytic domains of M1 peptidase family members including aminopeptidase N (APN) and leukotriene A4 hydrolase (LTA4H). All peptidases in this family bind a single catalytic zinc ion which is tetrahedrally co-ordinated by three amino acid ligands and a water molecule that forms the nucleophile upon activation during catalysis. APN preferentially cleaves neutral amino acids from the N-terminus of oligopeptides and is present in a variety of human tissues and cell types. APN expression is dysregulated in many inflammatory diseases and is enhanced in numerous tumor cells, making it a lead target in the development of anti-cancer and anti-inflammatory drugs. LTA4H is a bifunctional enzyme, possessing an aminopeptidase as well as an epoxide hydrolase activity. The two activities occupy different, but overlapping sites. The activity and physiological relevance of the aminopeptidase in LTA4H is as yet unknown, while the epoxide hydrolase converts leukotriene A4 (LTA4) into leukotriene B4 (LTB4), a potent chemotaxin that is fundamental to the inflammatory response of mammals. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSL66615.1|GT22 | 0.0 | 1 | 1331 | 1 | 1331 |
| QNP98605.1|GT22 | 2.09e-109 | 25 | 493 | 30 | 503 |
| AOW03560.1|GT22 | 2.89e-109 | 25 | 493 | 30 | 503 |
| CAG80582.1|GT22 | 2.89e-109 | 25 | 493 | 30 | 503 |
| CDR42052.1|GT22 | 2.04e-105 | 17 | 493 | 33 | 523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KX7_A | 6.95e-118 | 541 | 1324 | 88 | 877 | Crystal structure of human aminopeptidase A [Homo sapiens],4KX8_A Crystal structure of human aminopeptidase A complexed with amastatin [Homo sapiens],4KX9_A Crystal structure of human aminopeptidase A complexed with arginine [Homo sapiens],4KXA_A Crystal structure of human aminopeptidase A complexed with aspartate and calcium [Homo sapiens],4KXB_A Crystal structure of human aminopeptidase A complexed with bestatin [Homo sapiens],4KXC_A Crystal structure of human aminopeptidase A complexed with glutamate [Homo sapiens],4KXD_A Crystal structure of human aminopeptidase A complexed with glutamate and calcium [Homo sapiens] |
| 5MJ6_A | 2.72e-108 | 554 | 1323 | 95 | 872 | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. [Homo sapiens],5MJ6_B Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. [Homo sapiens] |
| 4P8Q_A | 4.22e-108 | 554 | 1322 | 96 | 872 | Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site [Homo sapiens],4P8Q_B Crystal Structure of Human Insulin Regulated Aminopeptidase with Alanine in Active Site [Homo sapiens],4PJ6_A Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site [Homo sapiens],4PJ6_B Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site [Homo sapiens] |
| 5C97_A | 5.37e-108 | 554 | 1323 | 126 | 903 | Insulin regulated aminopeptidase [Homo sapiens],5C97_B Insulin regulated aminopeptidase [Homo sapiens],6YDX_A Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor [Homo sapiens],6YDX_B Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor [Homo sapiens] |
| 4Z7I_A | 5.37e-108 | 554 | 1323 | 126 | 903 | Crystal structure of insulin regulated aminopeptidase in complex with ligand [Homo sapiens],4Z7I_B Crystal structure of insulin regulated aminopeptidase in complex with ligand [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9USX1|APE1_SCHPO | 1.09e-270 | 545 | 1328 | 89 | 882 | Aminopeptidase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ape1 PE=3 SV=1 |
| sp|Q59KZ1|APE2_CANAL | 1.17e-257 | 531 | 1327 | 126 | 922 | Aminopeptidase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=APE2 PE=1 SV=3 |
| sp|P32454|APE2_YEAST | 1.09e-250 | 524 | 1327 | 149 | 951 | Aminopeptidase 2, mitochondrial OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=APE2 PE=1 SV=4 |
| sp|P37898|AAP1_YEAST | 3.37e-242 | 533 | 1327 | 62 | 854 | Alanine/arginine aminopeptidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=AAP1 PE=1 SV=2 |
| sp|Q11011|PSA_MOUSE | 1.65e-165 | 534 | 1322 | 116 | 910 | Puromycin-sensitive aminopeptidase OS=Mus musculus OX=10090 GN=Npepps PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000004 | 0.000000 |
TMHMM Annotations download full data without filtering help
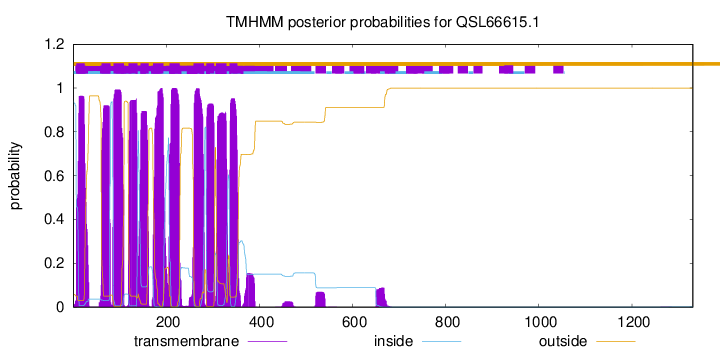
| Start | End |
|---|---|
| 7 | 25 |
| 61 | 80 |
| 87 | 109 |
| 119 | 138 |
| 145 | 162 |
| 172 | 194 |
| 207 | 229 |
| 259 | 281 |
| 286 | 303 |
| 307 | 329 |
| 336 | 353 |
