You are browsing environment: FUNGIDB
CAZyme Information: QSL66497.1
You are here: Home > Sequence: QSL66497.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis wakefieldiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis wakefieldiae | |||||||||||
| CAZyme ID | QSL66497.1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 151 | 387 | 1e-41 | 0.5578406169665809 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395127 | UQ_con | 1.87e-56 | 10 | 150 | 2 | 129 | Ubiquitin-conjugating enzyme. Proteins destined for proteasome-mediated degradation may be ubiquitinated. Ubiquitination follows conjugation of ubiquitin to a conserved cysteine residue of UBC homologs. TSG101 is one of several UBC homologs that lacks this active site cysteine. |
| 227410 | COG5078 | 1.21e-54 | 10 | 150 | 10 | 137 | Ubiquitin-protein ligase [Posttranslational modification, protein turnover, chaperones]. |
| 238117 | UBCc | 1.92e-50 | 10 | 150 | 4 | 131 | Ubiquitin-conjugating enzyme E2, catalytic (UBCc) domain. This is part of the ubiquitin-mediated protein degradation pathway in which a thiol-ester linkage forms between a conserved cysteine and the C-terminus of ubiquitin and complexes with ubiquitin protein ligase enzymes, E3. This pathway regulates many fundamental cellular processes. There are also other E2s which form thiol-ester linkages without the use of E3s as well as several UBC homologs (TSG101, Mms2, Croc-1 and similar proteins) which lack the active site cysteine essential for ubiquitination and appear to function in DNA repair pathways which were omitted from the scope of this CD. |
| 214562 | UBCc | 3.38e-49 | 10 | 149 | 2 | 129 | Ubiquitin-conjugating enzyme E2, catalytic domain homologues. Proteins destined for proteasome-mediated degradation may be ubiquitinated. Ubiquitination follows conjugation of ubiquitin to a conserved cysteine residue of UBC homologues. This pathway functions in regulating many fundamental processes required for cell viability.TSG101 is one of several UBC homologues that lacks this active site cysteine. |
| 177768 | PLN00172 | 1.25e-27 | 10 | 148 | 6 | 130 | ubiquitin conjugating enzyme; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 518 | 1 | 518 | |
| 1.45e-47 | 151 | 392 | 10 | 268 | |
| 1.45e-47 | 151 | 392 | 10 | 268 | |
| 1.45e-47 | 151 | 392 | 10 | 268 | |
| 2.01e-47 | 151 | 392 | 10 | 268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.19e-74 | 1 | 150 | 1 | 150 | Chain C, Ubiquitin-conjugating enzyme E2 15 [Schizosaccharomyces pombe 972h-],5KNL_F Chain F, Ubiquitin-conjugating enzyme E2 15 [Schizosaccharomyces pombe 972h-] |
|
| 8.01e-70 | 1 | 148 | 2 | 149 | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 [Homo sapiens],6D68_B Ube2G1 in complex with ubiquitin variant Ubv.G1.1 [Homo sapiens] |
|
| 4.59e-69 | 9 | 148 | 21 | 160 | Structure of human Ubiquitin-conjugating enzyme E2 G1 [Homo sapiens] |
|
| 1.82e-65 | 6 | 148 | 4 | 146 | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans [Caenorhabditis elegans] |
|
| 9.84e-51 | 10 | 161 | 8 | 159 | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g [Homo sapiens],3H8K_A Chain A, Ubiquitin-conjugating enzyme E2 G2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.44e-73 | 1 | 150 | 1 | 150 | Ubiquitin-conjugating enzyme E2 15 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ubc15 PE=1 SV=1 |
|
| 3.99e-69 | 1 | 148 | 1 | 148 | Ubiquitin-conjugating enzyme E2 G1 OS=Rattus norvegicus OX=10116 GN=Ube2g1 PE=2 SV=3 |
|
| 3.99e-69 | 1 | 148 | 1 | 148 | Ubiquitin-conjugating enzyme E2 G1 OS=Mus musculus OX=10090 GN=Ube2g1 PE=1 SV=3 |
|
| 3.99e-69 | 1 | 148 | 1 | 148 | Ubiquitin-conjugating enzyme E2 G1 OS=Homo sapiens OX=9606 GN=UBE2G1 PE=1 SV=3 |
|
| 4.38e-68 | 1 | 148 | 1 | 148 | Ubiquitin-conjugating enzyme E2 G1 OS=Macaca fascicularis OX=9541 GN=UBE2G1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000059 | 0.000000 |
TMHMM Annotations download full data without filtering help
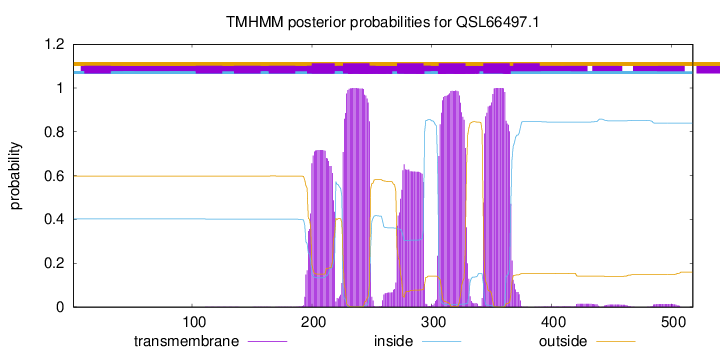
| Start | End |
|---|---|
| 200 | 219 |
| 226 | 248 |
| 271 | 293 |
| 306 | 328 |
| 343 | 365 |
