You are browsing environment: FUNGIDB
CAZyme Information: QSL64261.1
You are here: Home > Sequence: QSL64261.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis wakefieldiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis wakefieldiae | |||||||||||
| CAZyme ID | QSL64261.1 | |||||||||||
| CAZy Family | CBM48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.258:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT58 | 679 | 1037 | 5.7e-130 | 0.9862637362637363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 236327 | PRK08661 | 0.0 | 194 | 683 | 2 | 476 | prolyl-tRNA synthetase; Provisional |
| 398745 | ALG3 | 7.75e-176 | 679 | 1037 | 4 | 358 | ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. |
| 273062 | proS_fam_I | 3.13e-168 | 199 | 683 | 1 | 471 | prolyl-tRNA synthetase, family I. Prolyl-tRNA synthetase is a class II tRNA synthetase and is recognized by pfam model tRNA-synt_2b, which recognizes tRNA synthetases for Gly, His, Ser, and Pro. The prolyl-tRNA synthetases are divided into two widely divergent families. This family includes the archaeal enzyme, the Pro-specific domain of a human multifunctional tRNA ligase, and the enzyme from the spirochete Borrelia burgdorferi. The other family includes enzymes from Escherichia coli, Bacillus subtilis, Synechocystis PCC6803, and one of the two prolyL-tRNA synthetases of Saccharomyces cerevisiae. [Protein synthesis, tRNA aminoacylation] |
| 238401 | ProRS_core_arch_euk | 1.20e-140 | 205 | 451 | 1 | 261 | Prolyl-tRNA synthetase (ProRS) class II core catalytic domain. ProRS is a homodimer. It is responsible for the attachment of proline to the 3' OH group of ribose of the appropriate tRNA. This domain is primarily responsible for ATP-dependent formation of the enzyme bound aminoacyl-adenylate. Class II assignment is based upon its structure and the presence of three characteristic sequence motifs in the core domain. This subfamily contains the core domain of ProRS from archaea, the cytoplasm of eukaryotes and some bacteria. |
| 223519 | ProS | 5.98e-132 | 194 | 683 | 6 | 498 | Prolyl-tRNA synthetase [Translation, ribosomal structure and biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1055 | 1 | 1055 | |
| 7.24e-160 | 196 | 654 | 61 | 518 | |
| 1.47e-95 | 681 | 1046 | 39 | 405 | |
| 1.26e-91 | 677 | 1048 | 23 | 394 | |
| 1.38e-91 | 655 | 1046 | 21 | 405 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.61e-170 | 198 | 683 | 9 | 504 | Crystal Structure of Prolyl-tRNA Synthetase from Onchocerca volvulus with bound Halofuginone and nucleotide [Onchocerca volvulus] |
|
| 8.82e-167 | 189 | 683 | 7 | 511 | Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OSY_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OSZ_A Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OSZ_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT0_A Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT0_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT1_A Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT1_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT2_A Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT2_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT3_A Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens],7OT3_B Chain B, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens] |
|
| 9.77e-167 | 189 | 683 | 10 | 514 | Chain A, Bifunctional glutamate/proline--tRNA ligase [Homo sapiens] |
|
| 1.01e-166 | 189 | 683 | 11 | 515 | Crystal structure of human Prolyl-tRNA synthetase (PRS) in complex with inhibitor [Homo sapiens],5VAD_B Crystal structure of human Prolyl-tRNA synthetase (PRS) in complex with inhibitor [Homo sapiens] |
|
| 1.92e-166 | 189 | 683 | 30 | 534 | Crystal structure of human prolyl-tRNA synthetase (apo form) [Homo sapiens],4K87_A Crystal structure of human prolyl-tRNA synthetase (substrate bound form) [Homo sapiens],4K88_A Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.32e-234 | 1 | 683 | 1 | 715 | Putative proline--tRNA ligase C19C7.06 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=prs1 PE=3 SV=1 |
|
| 3.11e-197 | 66 | 683 | 42 | 687 | Putative proline--tRNA ligase YHR020W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YHR020W PE=1 SV=1 |
|
| 4.42e-169 | 173 | 683 | 31 | 529 | Proline--tRNA ligase, cytoplasmic OS=Arabidopsis thaliana OX=3702 GN=At3g62120 PE=1 SV=1 |
|
| 2.73e-165 | 164 | 683 | 1183 | 1713 | Bifunctional glutamate/proline--tRNA ligase OS=Drosophila melanogaster OX=7227 GN=GluProRS PE=1 SV=2 |
|
| 3.93e-160 | 197 | 683 | 7 | 500 | Proline--tRNA ligase OS=Encephalitozoon cuniculi (strain GB-M1) OX=284813 GN=ECU02_1360 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
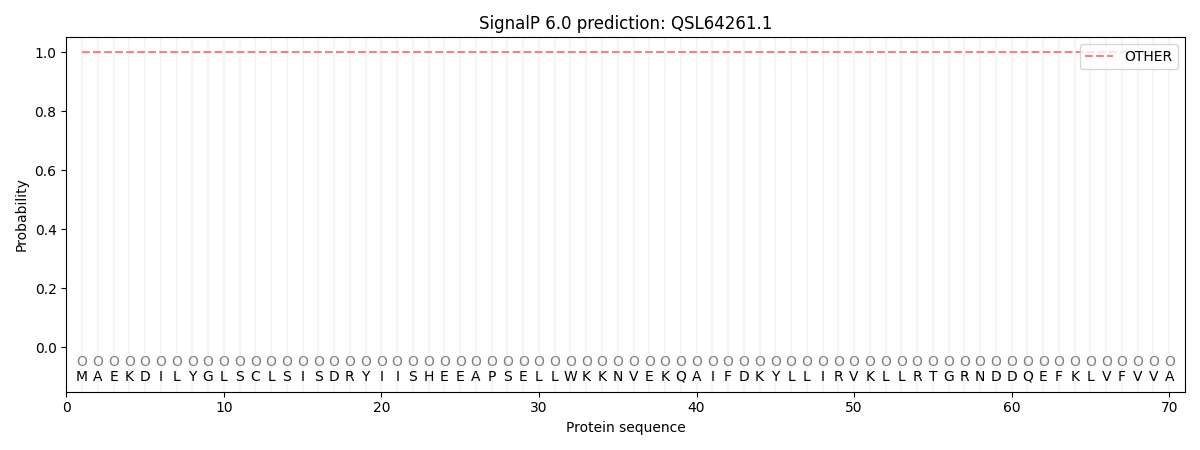
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999902 | 0.000129 |
TMHMM Annotations download full data without filtering help
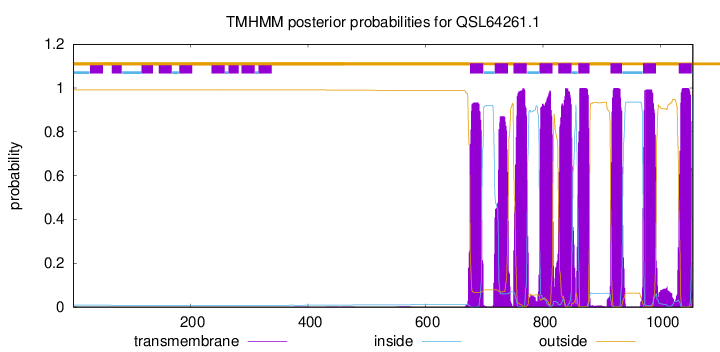
| Start | End |
|---|---|
| 676 | 698 |
| 718 | 740 |
| 750 | 772 |
| 794 | 816 |
| 826 | 848 |
| 860 | 879 |
| 915 | 934 |
| 970 | 992 |
| 1031 | 1053 |
