You are browsing environment: FUNGIDB
CAZyme Information: QRD85212.1
You are here: Home > Sequence: QRD85212.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus flavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus flavus | |||||||||||
| CAZyme ID | QRD85212.1 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | putative GPI anchored endo-1,3(4)-beta-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.39:3 | 3.2.1.-:1 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 71 | 305 | 8.5e-85 | 0.982532751091703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185690 | GH16_fungal_Lam16A_glucanase | 9.68e-144 | 31 | 326 | 2 | 293 | fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Group of fungal 1,3(4)-beta-D-glucanases, similar to Phanerochaete chrysosporium laminarinase 16A. Lam16A belongs to the 'nonspecific' 1,3(4)-beta-glucanase subfamily, although beta-1,6 branching and beta-1,4 bonds specifically define where Lam16A hydrolyzes its substrates, like curdlan (beta-1,3-glucan), lichenin (beta-1,3-1,4-mixed linkage glucan), and laminarin (beta-1,6-branched-1,3-glucan). |
| 185693 | GH16_laminarinase_like | 7.79e-12 | 126 | 281 | 92 | 215 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 223021 | PHA03247 | 4.96e-08 | 365 | 580 | 2736 | 2944 | large tegument protein UL36; Provisional |
| 411345 | gliding_GltJ | 8.40e-08 | 351 | 580 | 296 | 513 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 223021 | PHA03247 | 1.09e-07 | 327 | 589 | 2717 | 2987 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 667 | 1 | 667 | |
| 0.0 | 1 | 667 | 1 | 667 | |
| 0.0 | 1 | 667 | 1 | 667 | |
| 0.0 | 1 | 667 | 1 | 667 | |
| 0.0 | 1 | 667 | 1 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.14e-118 | 31 | 325 | 3 | 297 | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase [Paecilomyces sp. 'thermophila'],5JVV_B Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase [Paecilomyces sp. 'thermophila'] |
|
| 6.09e-78 | 31 | 325 | 2 | 295 | The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_B The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_C The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'],3WDU_D The complex structure of PtLic16A with cellobiose [Paecilomyces sp. 'thermophila'] |
|
| 6.28e-78 | 31 | 325 | 3 | 296 | The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_B The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_C The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDT_D The apo-form structure of PtLic16A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDV_A The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_B The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_C The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDV_D The complex structure of PtLic16A with cellotetraose [Paecilomyces sp. 'thermophila'] |
|
| 4.49e-77 | 31 | 325 | 1 | 294 | The complex structure of E113A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDY_B The complex structure of E113A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDY_C The complex structure of E113A with cellotetraose [Paecilomyces sp. 'thermophila'],3WDY_D The complex structure of E113A with cellotetraose [Paecilomyces sp. 'thermophila'] |
|
| 4.78e-77 | 31 | 325 | 3 | 296 | The apo-form structure of E113A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDW_B The apo-form structure of E113A from Paecilomyces thermophila [Paecilomyces sp. 'thermophila'],3WDX_A The complex structure of E113A with glucotriose [Paecilomyces sp. 'thermophila'],3WDX_B The complex structure of E113A with glucotriose [Paecilomyces sp. 'thermophila'] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 667 | 1 | 667 | Probable endo-1,3(4)-beta-glucanase AFLA_105200 OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=AFLA_105200 PE=3 SV=1 |
|
| 0.0 | 1 | 667 | 1 | 667 | Probable endo-1,3(4)-beta-glucanase AO090023000083 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=AO090023000083 PE=3 SV=1 |
|
| 4.58e-175 | 1 | 370 | 1 | 366 | Probable endo-1,3(4)-beta-glucanase AFUB_029980 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_029980 PE=3 SV=1 |
|
| 4.58e-175 | 1 | 370 | 1 | 366 | Probable endo-1,3(4)-beta-glucanase AFUA_2G14360 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_2G14360 PE=3 SV=1 |
|
| 1.00e-173 | 1 | 340 | 1 | 341 | Probable endo-1,3(4)-beta-glucanase NFIA_089530 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_089530 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
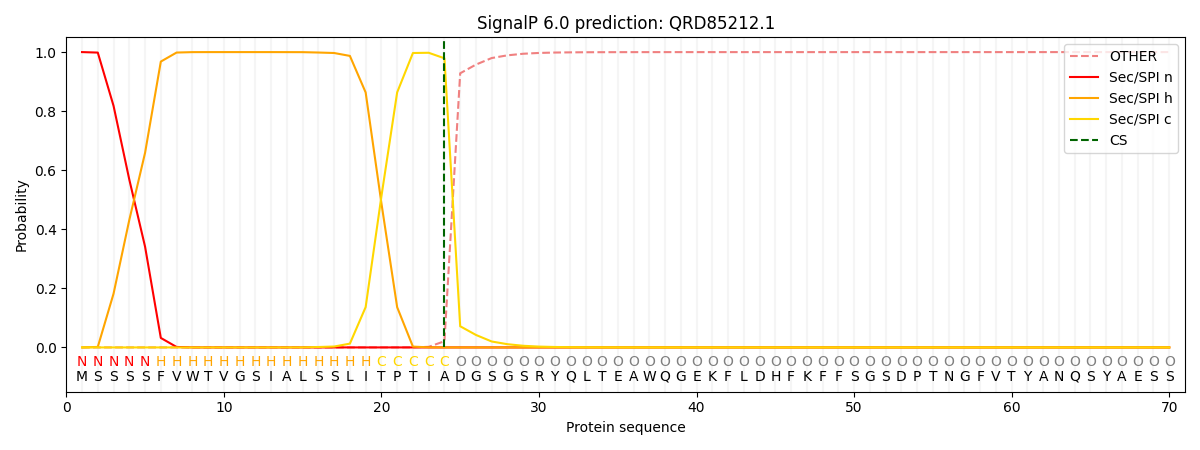
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999735 | CS pos: 24-25. Pr: 0.9790 |
