You are browsing environment: FUNGIDB
CAZyme Information: QRD83422.1
You are here: Home > Sequence: QRD83422.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus flavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus flavus | |||||||||||
| CAZyme ID | QRD83422.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | FMN-dependent dehydrogenase-domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 454 | 558 | 1.7e-20 | 0.7846153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200563 | CE4_HpPgdA_like | 6.05e-128 | 417 | 696 | 2 | 258 | Catalytic domain of Helicobacter pylori peptidoglycan deacetylase (HpPgdA) and similar proteins. This family is represented by a peptidoglycan deacetylase (HP0310, HpPgdA) from the gram-negative pathogen Helicobacter pylori. HpPgdA has the ability to bind a metal ion at the active site and is responsible for a peptidoglycan modification that counteracts the host immune response. It functions as a homotetramer. The monomer is composed of a 7-stranded barrel with detectable sequence similarity to the 6-stranded barrel NodB homology domain of polysaccharide deacetylase (DCA)-like proteins in the CE4 superfamily, which removes N-linked or O-linked acetyl groups from cell wall polysaccharides. In contrast to typical NodB-like DCAs, HpPgdA does not exhibit a solvent-accessible polysaccharide binding groove, suggesting that the enzyme binds a small molecule at the active site. |
| 239238 | FCB2_FMN | 6.82e-126 | 104 | 377 | 1 | 282 | Flavocytochrome b2 (FCB2) FMN-binding domain. FCB2 (AKA L-lactate:cytochrome c oxidoreductase) is a respiratory enzyme located in the intermembrane space of fungal mitochondria which catalyzes the oxidation of L-lactate to pyruvate. FCB2 also participates in a short electron-transport chain involving cytochrome c and cytochrome oxidase which ultimately directs the reducing equivalents gained from L-lactate oxidation to oxygen, yielding one molecule of ATP for every L-lactate molecule consumed. FCB2 is composed of 2 domains: a C-terminal flavin-binding domain, which includes the active site for lacate oxidation, and an N-terminal b2-cytochrome domain, required for efficient cytochrome c reduction. FCB2 is a homotetramer and contains two noncovalently bound cofactors, FMN and heme per subunit. |
| 395850 | FMN_dh | 8.30e-83 | 110 | 377 | 1 | 284 | FMN-dependent dehydrogenase. |
| 239203 | alpha_hydroxyacid_oxid_FMN | 3.18e-65 | 104 | 377 | 1 | 238 | Family of homologous FMN-dependent alpha-hydroxyacid oxidizing enzymes. This family occurs in both prokaryotes and eukaryotes. Members of this family include flavocytochrome b2 (FCB2), glycolate oxidase (GOX), lactate monooxygenase (LMO), mandelate dehydrogenase (MDH), and long chain hydroxyacid oxidase (LCHAO). In green plants, glycolate oxidase is one of the key enzymes in photorespiration where it oxidizes glycolate to glyoxylate. LMO catalyzes the oxidation of L-lactate to acetate and carbon dioxide. MDH oxidizes (S)-mandelate to phenylglyoxalate. It is an enzyme in the mandelate pathway that occurs in several strains of Pseudomonas which converts (R)-mandelate to benzoate. |
| 213021 | CE4_PuuE_HpPgdA_like | 1.29e-58 | 417 | 690 | 2 | 243 | Catalytic domain of bacterial PuuE allantoinases, Helicobacter pylori peptidoglycan deacetylase (HpPgdA), and similar proteins. This family is a member of the very large and functionally diverse carbohydrate esterase 4 (CE4) superfamily. It contains bacterial PuuE (purine utilization E) allantoinases, a peptidoglycan deacetylase from Helicobacter pylori (HpPgdA), Escherichia coli ArnD, and many uncharacterized homologs from all three kingdoms of life. PuuE allantoinase appears to be metal-independent and specifically catalyzes the hydrolysis of (S)-allantoin into allantoic acid. Different from PuuE allantoinase, HpPgdA has the ability to bind a metal ion at the active site and is responsible for a peptidoglycan modification that counteracts the host immune response. Both PuuE allantoinase and HpPgdA function as a homotetramer. The monomer is composed of a 7-stranded barrel with detectable sequence similarity to the 6-stranded barrel NodB homology domain of polysaccharide deacetylase (DCA)-like proteins in the CE4 superfamily, which removes N-linked or O-linked acetyl groups from cell wall polysaccharides. However, in contrast with the typical DCAs, PuuE allantoinase and HpPgdA might not exhibit a solvent-accessible polysaccharide binding groove and only recognize a small substrate molecule. ArnD catalyzes the deformylation of 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol to 4-amino-4-deoxy-L-arabinose-phosphoundecaprenol. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.21e-23 | 417 | 692 | 18 | 267 | |
| 5.30e-17 | 440 | 699 | 57 | 287 | |
| 7.14e-16 | 452 | 695 | 62 | 277 | |
| 7.97e-16 | 440 | 699 | 57 | 288 | |
| 1.08e-15 | 440 | 695 | 57 | 284 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.66e-138 | 405 | 699 | 29 | 322 | Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_B Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_C Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],3QBU_D Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori [Helicobacter pylori G27],4LY4_A Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_B Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_C Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27],4LY4_D Crystal structure of peptidoglycan deacetylase (HP0310) with Zinc from Helicobacter pylori [Helicobacter pylori G27] |
|
| 5.78e-70 | 1 | 377 | 6 | 414 | Chain A, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae],1SZE_B Chain B, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae] |
|
| 7.11e-70 | 1 | 377 | 1 | 409 | The 2.6 Angstroms Refined Structure Of The Escherichia Coli Recombinant Saccharomyces Cerevisiae Flavocytochrome B2-Sulphite Complex [Saccharomyces cerevisiae],1LTD_B The 2.6 Angstroms Refined Structure Of The Escherichia Coli Recombinant Saccharomyces Cerevisiae Flavocytochrome B2-Sulphite Complex [Saccharomyces cerevisiae] |
|
| 8.04e-70 | 1 | 377 | 6 | 414 | Molecular Structure Of Flavocytochrome B2 At 2.4 Angstroms Resolution [Saccharomyces cerevisiae],1FCB_B Molecular Structure Of Flavocytochrome B2 At 2.4 Angstroms Resolution [Saccharomyces cerevisiae],1KBI_A Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme [Saccharomyces cerevisiae],1KBI_B Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme [Saccharomyces cerevisiae] |
|
| 2.16e-69 | 1 | 377 | 6 | 414 | Chain A, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae],1SZF_B Chain B, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae],1SZG_A Chain A, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae],1SZG_B Chain B, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-155 | 1 | 377 | 1 | 409 | L-lactate dehydrogenase (cytochrome) OS=Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006) OX=441959 GN=TSTA_043820 PE=1 SV=1 |
|
| 2.67e-138 | 413 | 699 | 3 | 289 | Peptidoglycan deacetylase OS=Helicobacter pylori (strain G27) OX=563041 GN=pgdA PE=1 SV=1 |
|
| 4.80e-136 | 413 | 699 | 3 | 289 | Peptidoglycan deacetylase OS=Helicobacter pylori (strain ATCC 700392 / 26695) OX=85962 GN=pgdA PE=1 SV=1 |
|
| 1.75e-69 | 10 | 377 | 88 | 473 | L-lactate dehydrogenase (cytochrome) OS=Wickerhamomyces anomalus OX=4927 GN=CYB2 PE=1 SV=2 |
|
| 2.58e-68 | 1 | 377 | 86 | 494 | L-lactate dehydrogenase (cytochrome) OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CYB2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
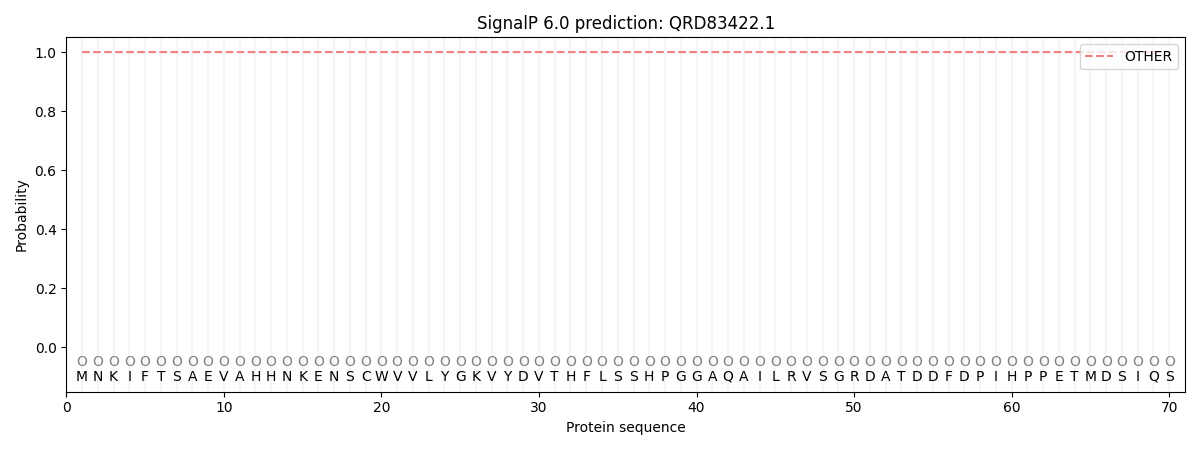
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000053 | 0.000000 |
