You are browsing environment: FUNGIDB
CAZyme Information: QRD83231.1
You are here: Home > Sequence: QRD83231.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
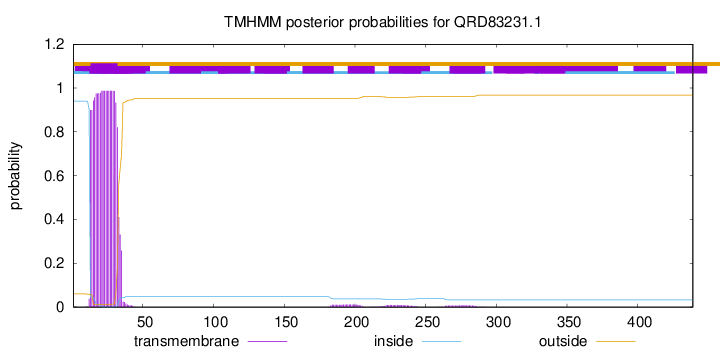
TMHMM annotations
Basic Information help
| Species | Aspergillus flavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus flavus | |||||||||||
| CAZyme ID | QRD83231.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238531 | PAN_AP_plant | 0.002 | 370 | 410 | 30 | 72 | Plant PAN/APPLE-like domain; present in plant S-receptor protein kinases and secreted glycoproteins. PAN/APPLE domains fulfill diverse biological functions by mediating protein-protein or protein-carbohydrate interactions. S-receptor protein kinases and S-locus glycoproteins are involved in sporophytic self-incompatibility response in Brassica, one of probably many molecular mechanisms, by which hermaphrodite flowering plants avoid self-fertilization. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 439 | 1 | 439 | |
| 0.0 | 1 | 439 | 1 | 439 | |
| 0.0 | 1 | 439 | 1 | 439 | |
| 0.0 | 1 | 439 | 1 | 439 | |
| 0.0 | 1 | 439 | 1 | 439 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999974 | 0.000027 |