You are browsing environment: FUNGIDB
CAZyme Information: QRD05983.1
You are here: Home > Sequence: QRD05983.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD05983.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | CBM20 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2NPU5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.55:5 | 1.14.99.55:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA13 | 18 | 247 | 5.4e-140 | 0.9956896551724138 |
| CBM20 | 289 | 379 | 1.2e-24 | 0.9666666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 412056 | AA13_LPMO-like | 2.50e-158 | 18 | 248 | 1 | 233 | AA13 lytic polysaccharide monooxygenase, and similar proteins. This family contains starch-degrading (also called starch-active) lytic polysaccharide monooxygenase (LPMO), a representative of the new CAZy AA13 family and classified as an auxiliary activity enzyme. This enzyme acts on alpha-linked glycosidic bonds and displays a binding surface that is quite different from those of LPMOs acting on beta-linked glycosidic bonds, indicating that the AA13 family proteins interact with their substrate in a distinct fashion. The active site contains an amino-terminal histidine-ligated mononuclear copper. This enzyme generates aldonic acid-terminated malto-oligosaccharides from retrograded starch and significantly boosts the conversion of this recalcitrant substrate to maltose by beta-amylase. |
| 99886 | CBM20_glucoamylase | 9.18e-52 | 282 | 388 | 1 | 106 | Glucoamylase (glucan1,4-alpha-glucosidase), C-terminal CBM20 (carbohydrate-binding module, family 20) domain. Glucoamylases are inverting, exo-acting starch hydrolases that hydrolyze starch and related polysaccharides by releasing the nonreducing end glucose. They are mainly active on alpha-1,4-glycosidic bonds but also have some activity towards 1,6-glycosidic bonds occurring in natural oligosaccharides. The ability of glucoamylases to cleave 1-6-glycosidic binds is called "debranching activity" and is of importance in industrial applications, where complete degradation of starch to glucose is needed. Most glucoamylases are multidomain proteins containing an N-terminal catalytic domain, a C-terminal CBM20 domain, and a highly O-glycosylated linker region that connects the two. The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 395557 | CBM_20 | 1.36e-38 | 289 | 384 | 1 | 95 | Starch binding domain. |
| 99883 | CBM20_alpha_amylase | 1.85e-27 | 289 | 389 | 1 | 95 | Alpha-amylase, C-terminal CBM20 (carbohydrate-binding module, family 20) domain. This domain is found in several bacterial and fungal alpha-amylases including the maltopentaose-forming amylases (G5-amylases). Most alpha-amylases have, in addition to the C-terminal CBM20 domain, an N-terminal catalytic domain belonging to glycosyl hydrolase family 13, which hydrolyzes internal alpha-1,4-glucosidic bonds in starch and related saccharides, yielding maltotriose and maltose. Two types of soluble substrates are used by alpha-amylases including long substrates (e.g. amylose) and short substrates (e.g. maltodextrins or maltooligosaccharides). The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 215006 | CBM_2 | 3.16e-25 | 289 | 375 | 1 | 87 | Starch binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.24e-286 | 1 | 389 | 1 | 389 | |
| 3.67e-224 | 8 | 389 | 8 | 393 | |
| 1.11e-216 | 1 | 388 | 1 | 403 | |
| 2.75e-213 | 1 | 389 | 1 | 378 | |
| 2.86e-188 | 1 | 389 | 1 | 353 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.07e-126 | 19 | 247 | 2 | 232 | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae [Aspergillus oryzae RIB40],5LSV_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7J_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7K_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],5T7N_A X-ray crystal structure of AA13 LPMO [Aspergillus oryzae RIB40],6TBQ_A AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae partially in Cu(II) state [Aspergillus oryzae RIB40],6TBR_A Glycosylated AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae in P1 space group [Aspergillus oryzae RIB40],6TBR_B Glycosylated AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae in P1 space group [Aspergillus oryzae RIB40],6TC4_A AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae measured with SSX [Aspergillus oryzae RIB40] |
|
| 5.04e-28 | 282 | 389 | 1 | 108 | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, MINIMIZED AVERAGE STRUCTURE [Aspergillus niger],1ACZ_A GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, 5 STRUCTURES [Aspergillus niger],1KUL_A Chain A, GLUCOAMYLASE [Aspergillus niger],1KUM_A Chain A, GLUCOAMYLASE [Aspergillus niger],5GHL_A Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_B Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_C Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_D Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger] |
|
| 1.48e-22 | 292 | 389 | 518 | 616 | Structure of the catalytic domain of Aspergillus niger Glucoamylase [Aspergillus niger] |
|
| 1.01e-17 | 230 | 375 | 441 | 583 | Chain A, GLUCOAMYLASE [Trichoderma reesei],2VN7_A Chain A, GLUCOAMYLASE [Trichoderma reesei] |
|
| 1.37e-10 | 289 | 388 | 507 | 606 | Chain A, Glucoamylase P [Amorphotheca resinae],6FHW_B Chain B, Glucoamylase P [Amorphotheca resinae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.15e-24 | 289 | 389 | 538 | 639 | Glucoamylase I OS=Aspergillus kawachii OX=1069201 GN=gaI PE=1 SV=1 |
|
| 1.97e-24 | 275 | 389 | 499 | 612 | Glucoamylase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=glaA PE=2 SV=2 |
|
| 9.25e-24 | 289 | 389 | 538 | 639 | Glucoamylase OS=Aspergillus usamii OX=186680 GN=glaA PE=3 SV=1 |
|
| 5.37e-23 | 280 | 389 | 518 | 626 | Glucoamylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gla-1 PE=1 SV=3 |
|
| 7.91e-22 | 292 | 389 | 542 | 640 | Glucoamylase OS=Aspergillus niger OX=5061 GN=GLAA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
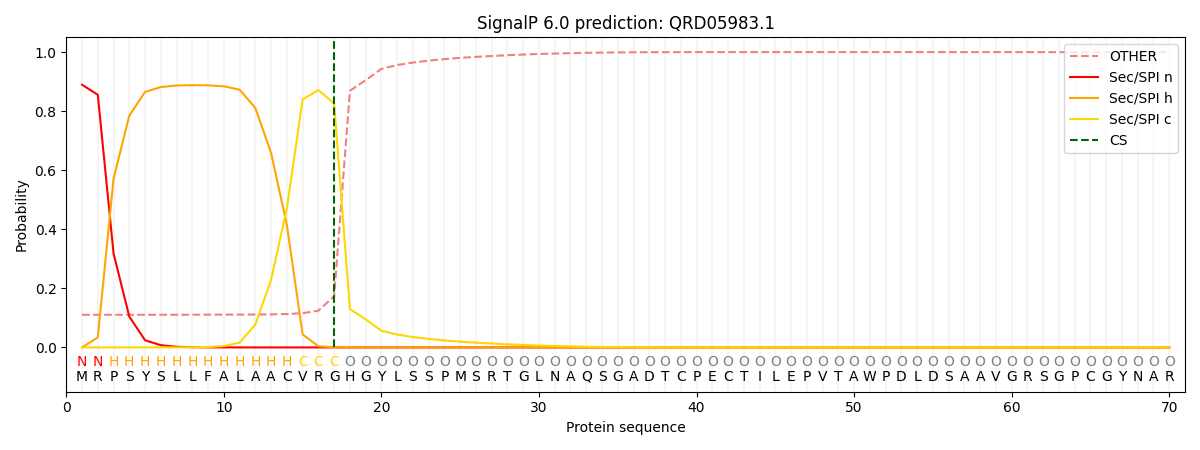
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.123889 | 0.876111 | CS pos: 17-18. Pr: 0.8258 |
