You are browsing environment: FUNGIDB
CAZyme Information: QRD05934.1
You are here: Home > Sequence: QRD05934.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD05934.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Cellulase [Source:UniProtKB/TrEMBL;Acc:A0A7U2I9Y7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:59 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 28 | 225 | 4.1e-72 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 3.18e-64 | 27 | 226 | 1 | 214 | Glycosyl hydrolase family 45. |
| 395595 | CBM_1 | 5.74e-08 | 394 | 422 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.55e-07 | 394 | 426 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 197593 | fCBD | 1.09e-05 | 312 | 343 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 2.30e-05 | 312 | 339 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 428 | 1 | 428 | |
| 1.51e-192 | 1 | 342 | 1 | 343 | |
| 1.02e-93 | 5 | 287 | 3 | 299 | |
| 1.02e-93 | 5 | 287 | 3 | 299 | |
| 1.02e-93 | 5 | 287 | 3 | 299 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.07e-89 | 5 | 286 | 3 | 292 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 4.08e-82 | 25 | 230 | 2 | 205 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
|
| 4.66e-82 | 25 | 230 | 5 | 208 | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126] |
|
| 2.31e-81 | 28 | 230 | 3 | 204 | Endoglucanase from Humicola insolens AT 1.7A resolution [Humicola insolens] |
|
| 1.67e-80 | 28 | 230 | 3 | 204 | Chain A, ENDOGLUCANASE V CELLOHEXAOSE COMPLEX [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.70e-83 | 20 | 425 | 14 | 373 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 1.34e-79 | 28 | 230 | 3 | 204 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
|
| 1.93e-64 | 20 | 231 | 14 | 225 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 6.49e-43 | 1 | 225 | 1 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 2.48e-36 | 11 | 230 | 7 | 239 | Endoglucanase 1 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=EGL1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
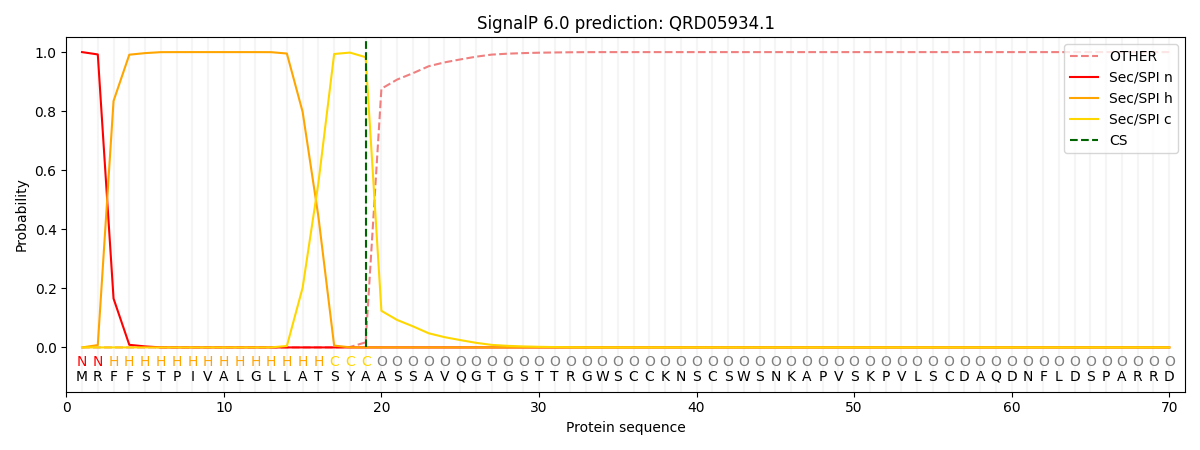
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000221 | 0.999768 | CS pos: 19-20. Pr: 0.9827 |
