You are browsing environment: FUNGIDB
CAZyme Information: QRD04149.1
You are here: Home > Sequence: QRD04149.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD04149.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Glycoside hydrolase [Source:UniProtKB/TrEMBL;Acc:A0A7U2FH85] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.38e-15 | 25 | 300 | 47 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 4.25e-04 | 206 | 310 | 190 | 309 | Glycosyl hydrolases family 17. |
| 223021 | PHA03247 | 0.005 | 327 | 491 | 2717 | 2876 | large tegument protein UL36; Provisional |
| 407229 | PARM | 0.009 | 328 | 413 | 88 | 180 | PARM. Human PARM-1 is a mucin-like, androgen-regulated transmembrane protein that is present in most tissues, with high levels in the heart, kidney and placenta. It has been shown to be induced and expressed in prostate after castration and may have a role in cell proliferation and immortalisation in prostate cancer. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 602 | 1 | 602 | |
| 6.56e-109 | 3 | 396 | 2 | 377 | |
| 8.83e-52 | 11 | 344 | 11 | 347 | |
| 5.70e-51 | 23 | 332 | 23 | 346 | |
| 1.04e-50 | 8 | 320 | 11 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.58e-12 | 23 | 272 | 37 | 254 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.64e-50 | 10 | 320 | 7 | 320 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglC PE=3 SV=1 |
|
| 2.08e-46 | 11 | 320 | 8 | 320 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglC PE=3 SV=1 |
|
| 2.65e-46 | 11 | 313 | 8 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=eglC PE=3 SV=1 |
|
| 2.88e-46 | 11 | 313 | 8 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=eglC PE=3 SV=1 |
|
| 2.95e-43 | 11 | 313 | 8 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
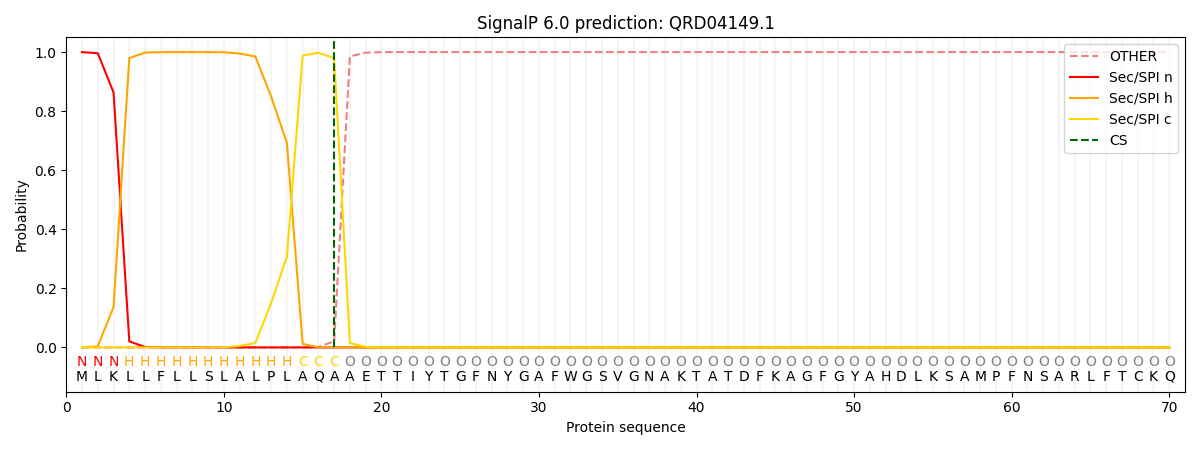
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000272 | 0.999709 | CS pos: 17-18. Pr: 0.9794 |
