You are browsing environment: FUNGIDB
CAZyme Information: QRD03005.1
You are here: Home > Sequence: QRD03005.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD03005.1 | |||||||||||
| CAZy Family | GH43|CBM91 | |||||||||||
| CAZyme Description | NAD(P)-binding protein [Source:UniProtKB/TrEMBL;Acc:Q0U243] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.49:6 | 3.2.1.52:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 9 | 167 | 1.6e-26 | 0.39598997493734334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223745 | MviM | 1.43e-37 | 5 | 224 | 1 | 213 | Predicted dehydrogenase [General function prediction only]. |
| 396129 | GFO_IDH_MocA | 7.86e-17 | 8 | 136 | 1 | 120 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| 397161 | GFO_IDH_MocA_C | 3.25e-14 | 149 | 423 | 2 | 200 | Oxidoreductase family, C-terminal alpha/beta domain. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| 182305 | PRK10206 | 3.48e-06 | 69 | 178 | 55 | 156 | putative oxidoreductase; Provisional |
| 183212 | PRK11579 | 3.63e-04 | 9 | 190 | 6 | 173 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.05e-31 | 9 | 429 | 396 | 753 | |
| 6.23e-18 | 9 | 430 | 5 | 364 | |
| 9.60e-14 | 1 | 239 | 12 | 236 | |
| 1.43e-12 | 9 | 420 | 57 | 445 | |
| 3.94e-12 | 4 | 229 | 36 | 274 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.13e-11 | 65 | 215 | 60 | 207 | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A02_B Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A02_C Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A02_D Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A02_E Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A02_F Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol [Caulobacter vibrioides CB15],5A03_A Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A03_B Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A03_C Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A03_D Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A03_E Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A03_F Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose [Caulobacter vibrioides CB15],5A04_A Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A04_B Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A04_C Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A04_D Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A04_E Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A04_F Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose [Caulobacter vibrioides CB15],5A05_A Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A05_B Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A05_C Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A05_D Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A05_E Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A05_F Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose [Caulobacter vibrioides CB15],5A06_A Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15],5A06_B Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15],5A06_C Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15],5A06_D Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15],5A06_E Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15],5A06_F Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol [Caulobacter vibrioides CB15] |
|
| 2.70e-11 | 80 | 215 | 65 | 193 | Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_B Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_C Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_D Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021] |
|
| 1.50e-09 | 9 | 215 | 4 | 199 | The crystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 [Bordetella bronchiseptica] |
|
| 2.93e-09 | 70 | 198 | 74 | 203 | Crystal structure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8],1ZH8_B Crystal structure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8] |
|
| 1.01e-08 | 68 | 215 | 55 | 198 | CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua],3E18_B CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-75 | 8 | 431 | 2 | 423 | Putative oxidoreductase YteT OS=Bacillus subtilis (strain 168) OX=224308 GN=yteT PE=2 SV=1 |
|
| 5.76e-11 | 80 | 215 | 64 | 192 | Inositol 2-dehydrogenase OS=Rhizobium meliloti (strain 1021) OX=266834 GN=idhA PE=1 SV=2 |
|
| 1.13e-09 | 48 | 217 | 36 | 198 | Uncharacterized oxidoreductase YrbE OS=Bacillus subtilis (strain 168) OX=224308 GN=yrbE PE=3 SV=2 |
|
| 4.33e-08 | 30 | 202 | 25 | 194 | Oxidoreductase P35 OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22240 PE=2 SV=1 |
|
| 7.04e-08 | 9 | 193 | 76 | 267 | Glycosyl hydrolase family 109 protein 1 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0017 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
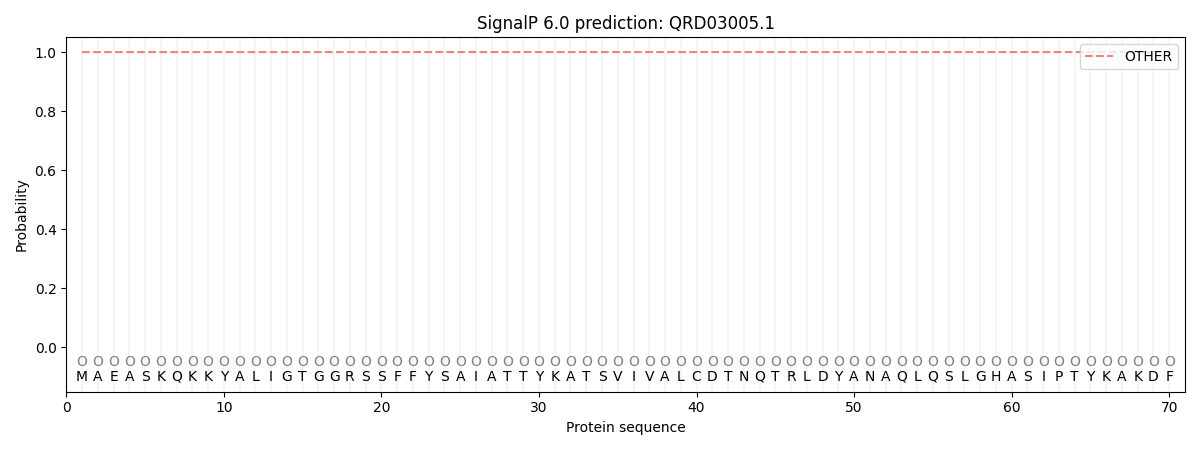
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000045 | 0.000000 |
