You are browsing environment: FUNGIDB
CAZyme Information: QRD02987.1
You are here: Home > Sequence: QRD02987.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD02987.1 | |||||||||||
| CAZy Family | GH43|CBM91 | |||||||||||
| CAZyme Description | Extracellular serine-rich protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2FCX1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE18 | 249 | 620 | 2.6e-189 | 0.9945504087193461 |
| CBM87 | 40 | 248 | 3e-90 | 0.9954128440366973 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 213023 | CE4_NodB_like_5s_6s | 3.18e-04 | 352 | 425 | 38 | 102 | Putative catalytic NodB homology domain of PgaB, IcaB, and similar proteins which consist of a deformed (beta/alpha)8 barrel fold with 5- or 6-strands. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes bacterial poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB, hemin storage system HmsF protein in gram-negative species, intercellular adhesion proteins IcaB, and many uncharacterized prokaryotic polysaccharide deacetylases. It also includes a putative polysaccharide deacetylase YxkH encoded by the Bacillus subtilis yxkH gene, which is one of six polysaccharide deacetylase gene homologs present in the Bacillus subtilis genome. Sequence comparison shows all family members contain a conserved domain similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, which consists of a deformed (beta/alpha)8 barrel fold with 6 or 7 strands. However, in this family, most proteins have 5 strands and some have 6 strands. Moreover, long insertions are found in many family members, whose function remains unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 693 | 1 | 693 | |
| 0.0 | 1 | 688 | 1 | 694 | |
| 0.0 | 7 | 693 | 5 | 687 | |
| 1.59e-249 | 29 | 690 | 39 | 705 | |
| 8.33e-238 | 32 | 693 | 25 | 691 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.88e-235 | 32 | 693 | 22 | 684 | Crystal structure of Agd3 a novel carbohydrate deacetylase [Aspergillus fumigatus Af293] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
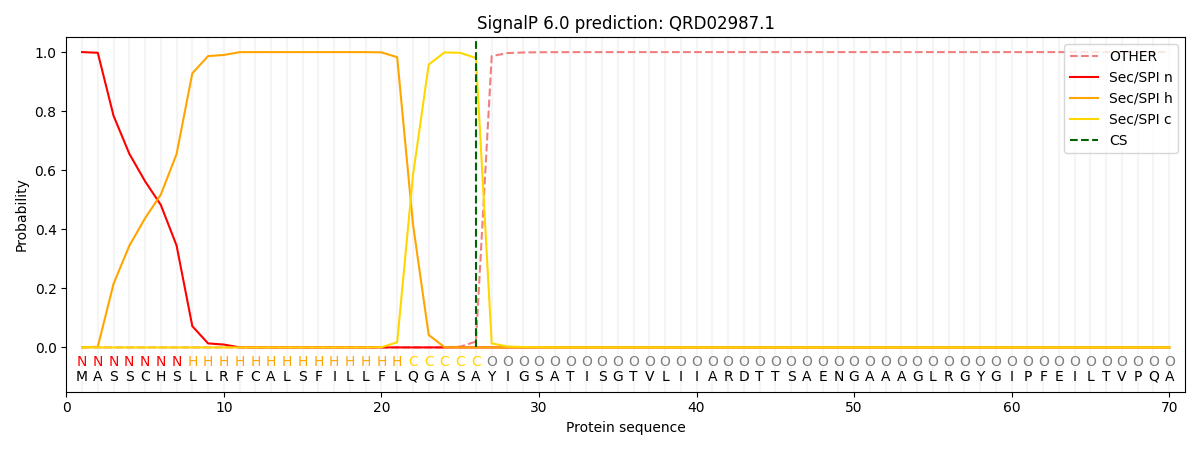
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999772 | CS pos: 26-27. Pr: 0.9801 |

