You are browsing environment: FUNGIDB
CAZyme Information: QRD02923.1
Basic Information
help
| Species |
Parastagonospora nodorum
|
| Lineage |
Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum
|
| CAZyme ID |
QRD02923.1
|
| CAZy Family |
GH43 |
| CAZyme Description |
Mannan endo-1,4-beta-mannosidase [Source:UniProtKB/TrEMBL;Acc:A0A7U2FDA8]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 361 |
|
39386.15 |
6.7397 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PnodorumSN15 |
17580 |
321614 |
132 |
17448
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
43 |
322 |
1.8e-160 |
0.9929328621908127 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 395098
|
Cellulase |
7.61e-09 |
77 |
316 |
51 |
268 |
Cellulase (glycosyl hydrolase family 5). |
| 225344
|
BglC |
3.27e-05 |
80 |
341 |
105 |
385 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 4.77e-269 |
1 |
361 |
1 |
361 |
| 4.77e-269 |
1 |
361 |
1 |
361 |
| 5.96e-206 |
1 |
361 |
1 |
362 |
| 1.27e-200 |
7 |
361 |
9 |
362 |
| 8.98e-194 |
19 |
361 |
20 |
362 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.20e-10 |
24 |
277 |
21 |
300 |
Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 7.46e-09 |
44 |
276 |
81 |
335 |
Mannan endo-1,4-beta-mannosidase 5 OS=Solanum lycopersicum OX=4081 GN=MAN5 PE=2 SV=1 |
| 7.46e-09 |
44 |
276 |
81 |
335 |
Mannan endo-1,4-beta-mannosidase 2 OS=Solanum lycopersicum OX=4081 GN=MAN2 PE=2 SV=2 |
This protein is predicted as SP
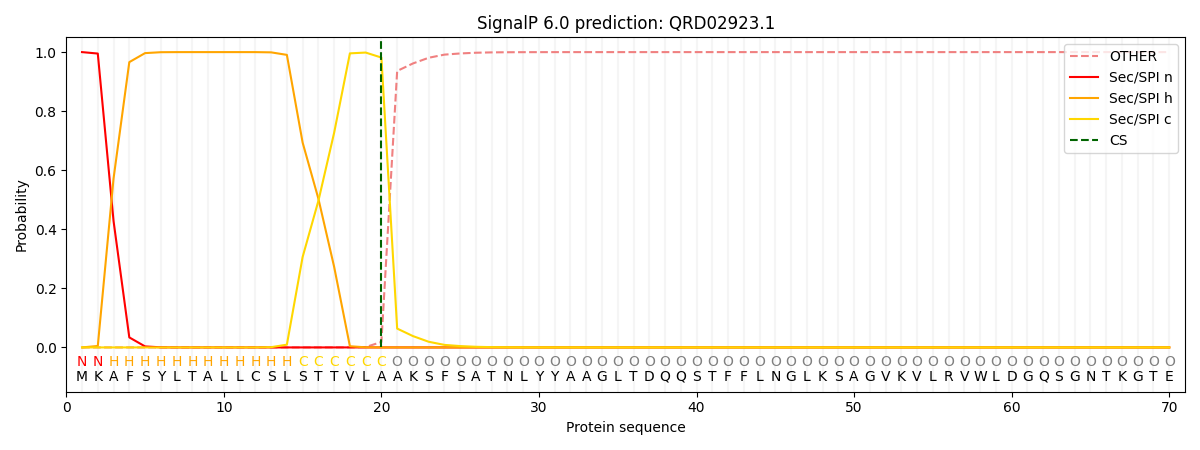
| Other |
SP_Sec_SPI |
CS Position |
| 0.000255 |
0.999724 |
CS pos: 20-21. Pr: 0.9815 |
There is no transmembrane helices in QRD02923.1.

