You are browsing environment: FUNGIDB
CAZyme Information: QRD02548.1
You are here: Home > Sequence: QRD02548.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD02548.1 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | cellulase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:59 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 26 | 225 | 2.1e-92 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 1.41e-88 | 25 | 225 | 1 | 213 | Glycosyl hydrolase family 45. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.76e-183 | 1 | 233 | 1 | 233 | |
| 4.49e-162 | 1 | 233 | 1 | 233 | |
| 1.27e-145 | 1 | 230 | 1 | 229 | |
| 8.81e-138 | 1 | 227 | 1 | 226 | |
| 1.99e-122 | 6 | 233 | 2 | 235 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.71e-107 | 19 | 228 | 19 | 224 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 2.27e-103 | 26 | 228 | 3 | 202 | Endoglucanase from Humicola insolens AT 1.7A resolution [Humicola insolens] |
|
| 4.80e-102 | 26 | 228 | 3 | 202 | Endoglucanase V [Humicola insolens] |
|
| 5.32e-102 | 26 | 228 | 3 | 202 | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX [Humicola insolens] |
|
| 5.32e-102 | 25 | 228 | 4 | 203 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.05e-109 | 10 | 228 | 7 | 222 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 2.74e-101 | 26 | 228 | 3 | 202 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
|
| 2.34e-93 | 9 | 227 | 4 | 221 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 9.32e-57 | 26 | 225 | 40 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 1.20e-49 | 26 | 228 | 269 | 501 | Endoglucanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
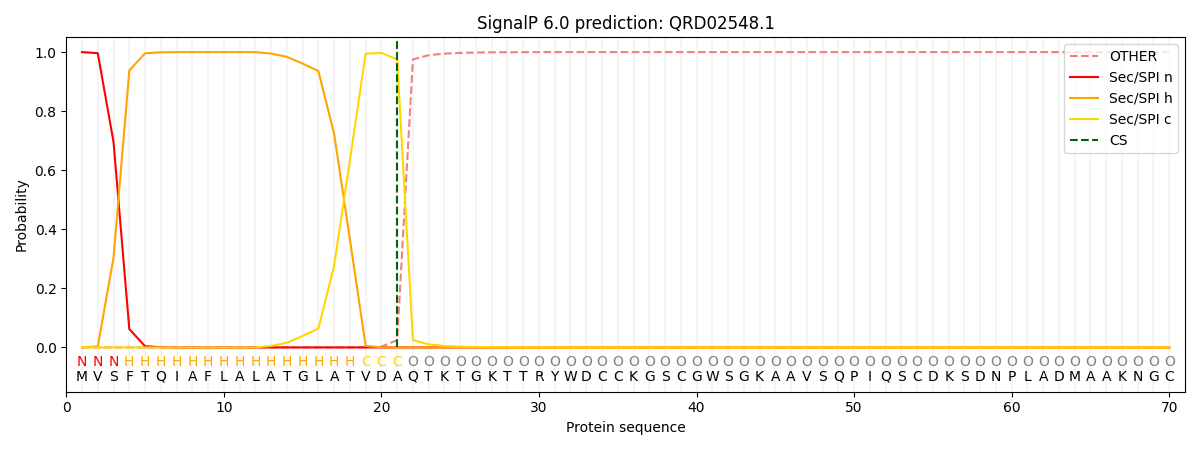
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000243 | 0.999732 | CS pos: 21-22. Pr: 0.9753 |
