You are browsing environment: FUNGIDB
CAZyme Information: QRD01685.1
You are here: Home > Sequence: QRD01685.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD01685.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Glycoside hydrolase [Source:UniProtKB/TrEMBL;Acc:A0A7U2I4M4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 209 | 457 | 4e-22 | 0.9421221864951769 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 8.06e-56 | 195 | 456 | 42 | 301 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 2.26e-06 | 377 | 457 | 216 | 298 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.12e-283 | 1 | 457 | 1 | 457 | |
| 4.47e-125 | 1 | 457 | 1 | 440 | |
| 4.23e-120 | 1 | 456 | 1 | 439 | |
| 5.20e-62 | 200 | 456 | 119 | 364 | |
| 1.03e-61 | 200 | 456 | 119 | 364 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-55 | 200 | 456 | 135 | 383 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 4.21e-55 | 185 | 455 | 115 | 375 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 1.60e-53 | 200 | 456 | 138 | 386 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 9.16e-38 | 184 | 456 | 336 | 598 | Probable beta-glucosidase btgE OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgE PE=3 SV=1 |
|
| 9.16e-38 | 184 | 456 | 336 | 598 | Probable beta-glucosidase btgE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
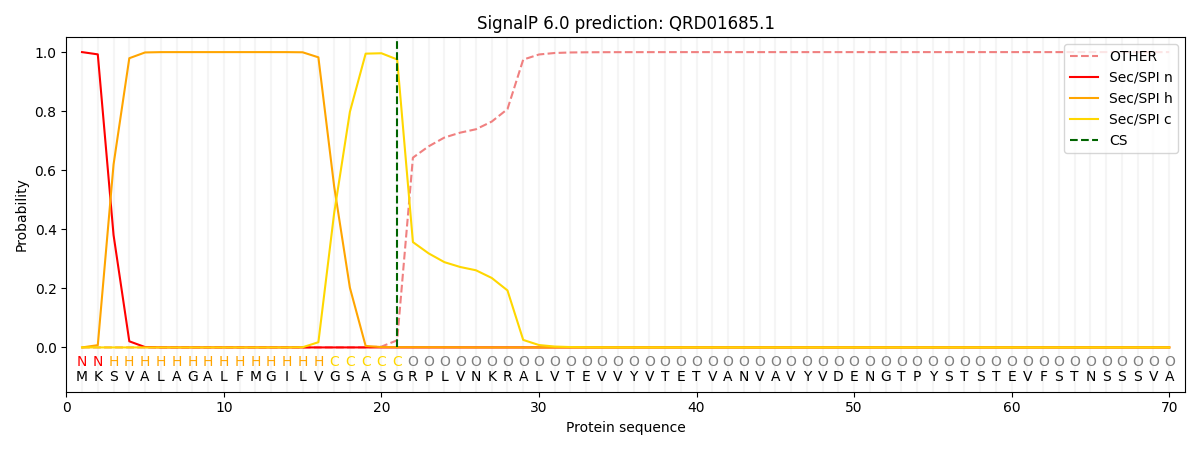
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000310 | 0.999663 | CS pos: 21-22. Pr: 0.9751 |
