You are browsing environment: FUNGIDB
CAZyme Information: QRD00841.1
You are here: Home > Sequence: QRD00841.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRD00841.1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | feruloyl esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:1 | 3.2.1.8:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 55 | 267 | 4.3e-20 | 0.7268722466960352 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 1.08e-26 | 55 | 296 | 46 | 278 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 226584 | COG4099 | 3.92e-09 | 57 | 230 | 175 | 334 | Predicted peptidase [General function prediction only]. |
| 224423 | DAP2 | 7.73e-05 | 58 | 202 | 380 | 520 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.72e-17 | 132 | 222 | 224 | 317 | |
| 1.80e-16 | 32 | 297 | 167 | 425 | |
| 9.05e-16 | 32 | 281 | 14 | 228 | |
| 1.23e-14 | 27 | 240 | 236 | 425 | |
| 3.84e-14 | 29 | 311 | 12 | 261 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.32e-16 | 56 | 232 | 292 | 457 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
|
| 1.37e-11 | 56 | 278 | 58 | 245 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
|
| 1.71e-09 | 56 | 232 | 45 | 202 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
|
| 6.45e-09 | 13 | 222 | 15 | 205 | Feruloyl esterase D OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=faeD-3.544 PE=1 SV=1 |
|
| 7.48e-09 | 14 | 268 | 7 | 245 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
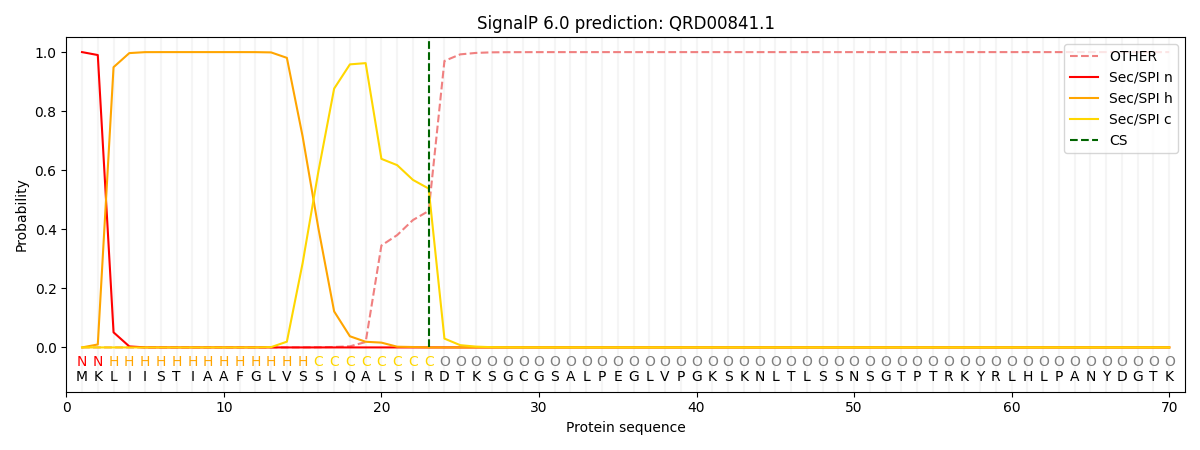
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000231 | 0.999751 | CS pos: 23-24. Pr: 0.5379 |
