You are browsing environment: FUNGIDB
CAZyme Information: QRC98595.1
You are here: Home > Sequence: QRC98595.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC98595.1 | |||||||||||
| CAZy Family | GH132 | |||||||||||
| CAZyme Description | Glycoside hydrolase [Source:UniProtKB/TrEMBL;Acc:A0A7U2I0C8] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 240 | 486 | 3.1e-20 | 0.9163987138263665 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 4.52e-57 | 194 | 483 | 20 | 299 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 223021 | PHA03247 | 3.14e-09 | 50 | 219 | 2688 | 2861 | large tegument protein UL36; Provisional |
| 411474 | fibronec_FbpA | 6.75e-07 | 37 | 195 | 149 | 306 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411490 | pneumo_PspA | 7.70e-07 | 106 | 204 | 354 | 434 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| 223039 | PHA03307 | 4.45e-06 | 37 | 222 | 46 | 250 | transcriptional regulator ICP4; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.77e-308 | 1 | 489 | 1 | 489 | |
| 2.61e-151 | 184 | 488 | 584 | 872 | |
| 1.14e-122 | 206 | 488 | 374 | 653 | |
| 1.23e-65 | 171 | 485 | 117 | 410 | |
| 1.04e-64 | 214 | 489 | 148 | 413 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-62 | 216 | 486 | 298 | 556 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 6.58e-59 | 223 | 488 | 311 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 6.58e-59 | 223 | 488 | 311 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 1.38e-58 | 218 | 488 | 293 | 551 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 2.43e-58 | 223 | 486 | 311 | 562 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
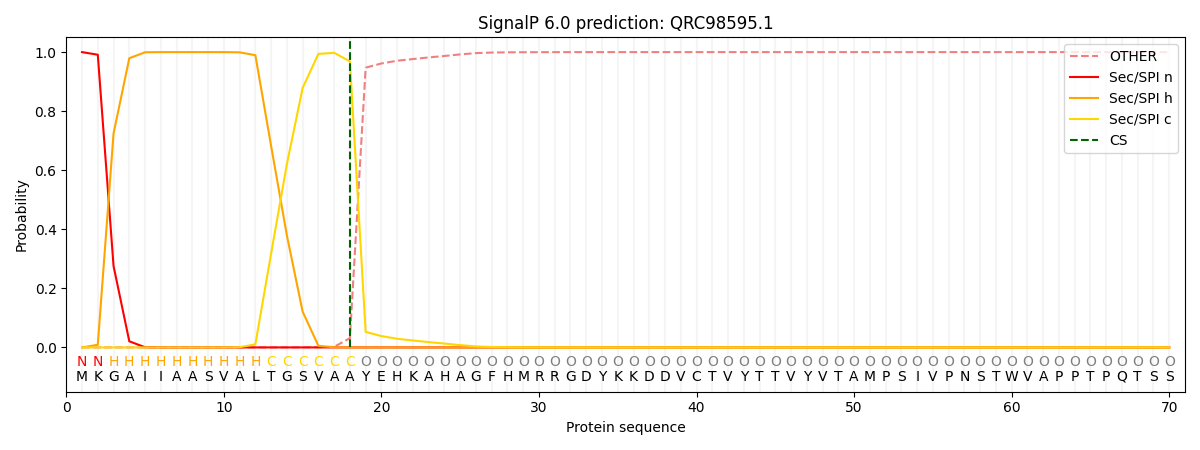
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000521 | 0.999472 | CS pos: 18-19. Pr: 0.9686 |
