You are browsing environment: FUNGIDB
CAZyme Information: QRC97346.1
You are here: Home > Sequence: QRC97346.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC97346.1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Carbohydrate esterase family 5 protein [Source:UniProtKB/TrEMBL;Acc:Q0UH92] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 29 | 232 | 1.6e-36 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 8.57e-16 | 29 | 233 | 1 | 173 | Cutinase. |
| 411474 | fibronec_FbpA | 7.99e-05 | 222 | 305 | 153 | 228 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411474 | fibronec_FbpA | 9.89e-05 | 242 | 307 | 196 | 259 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 411345 | gliding_GltJ | 1.99e-04 | 241 | 307 | 423 | 490 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 411474 | fibronec_FbpA | 2.52e-04 | 242 | 308 | 231 | 288 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.81e-207 | 1 | 324 | 1 | 324 | |
| 5.86e-114 | 4 | 253 | 5 | 254 | |
| 1.58e-102 | 4 | 244 | 4 | 242 | |
| 1.11e-92 | 7 | 233 | 13 | 243 | |
| 1.84e-59 | 27 | 238 | 22 | 235 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.25e-17 | 30 | 233 | 2 | 206 | Acetylxylan Esterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
|
| 8.25e-17 | 30 | 233 | 2 | 206 | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
|
| 8.25e-17 | 30 | 233 | 2 | 206 | Iodinated Complex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
|
| 3.09e-13 | 83 | 233 | 57 | 206 | Chain A, ACETYL XYLAN ESTERASE [Trichoderma reesei],1QOZ_B Chain B, ACETYL XYLAN ESTERASE [Trichoderma reesei] |
|
| 4.00e-06 | 30 | 233 | 17 | 193 | Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.92e-16 | 30 | 233 | 29 | 233 | Acetylxylan esterase 2 OS=Talaromyces purpureogenus OX=1266744 GN=axe-2 PE=1 SV=1 |
|
| 2.07e-12 | 29 | 233 | 32 | 237 | Acetylxylan esterase OS=Hypocrea jecorina OX=51453 GN=axe1 PE=1 SV=1 |
|
| 2.63e-12 | 83 | 234 | 80 | 230 | Cutinase 11 OS=Verticillium dahliae OX=27337 GN=VD0003_g7577 PE=1 SV=1 |
|
| 5.51e-10 | 12 | 233 | 29 | 226 | Phospholipase Culp4 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut4 PE=1 SV=3 |
|
| 1.50e-08 | 7 | 233 | 8 | 199 | Cutinase CUT1 OS=Coprinopsis cinerea OX=5346 GN=CUT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
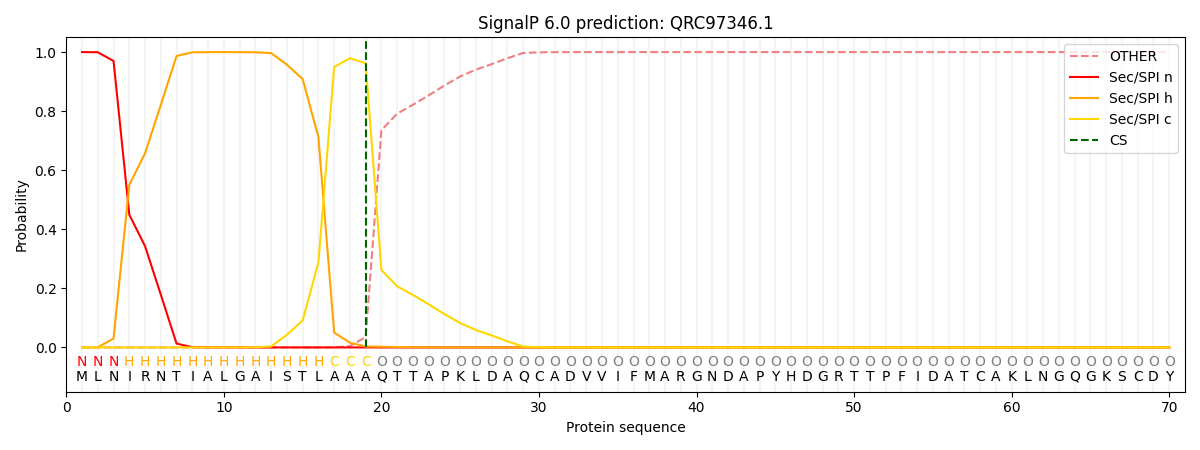
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000363 | 0.999624 | CS pos: 19-20. Pr: 0.9620 |
