You are browsing environment: FUNGIDB
QRC96084.1
Basic Information
help
Species
Parastagonospora nodorum
Lineage
Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum
CAZyme ID
QRC96084.1
CAZy Family
CE16
CAZyme Description
Proteophosphoglycan ppg4 [Source:UniProtKB/TrEMBL;Acc:A0A7U2EZP3]
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
430
46435.94
6.9118
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_PnodorumSN15
17580
321614
132
17448
Gene Location
No EC number prediction in QRC96084.1.
Family
Start
End
Evalue
family coverage
AA14
18
289
2.2e-79
0.9882352941176471
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
237865
PRK14951
3.10e-04
290
407
374
492
DNA polymerase III subunits gamma and tau; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
3.24e-263
1
430
1
430
8.12e-127
5
288
6
286
7.31e-125
1
288
1
285
1.07e-124
8
289
8
287
1.72e-124
8
292
8
289
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.85e-40
18
288
1
261
Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina],5NO7_B Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina]
more
QRC96084.1 has no Swissprot hit.
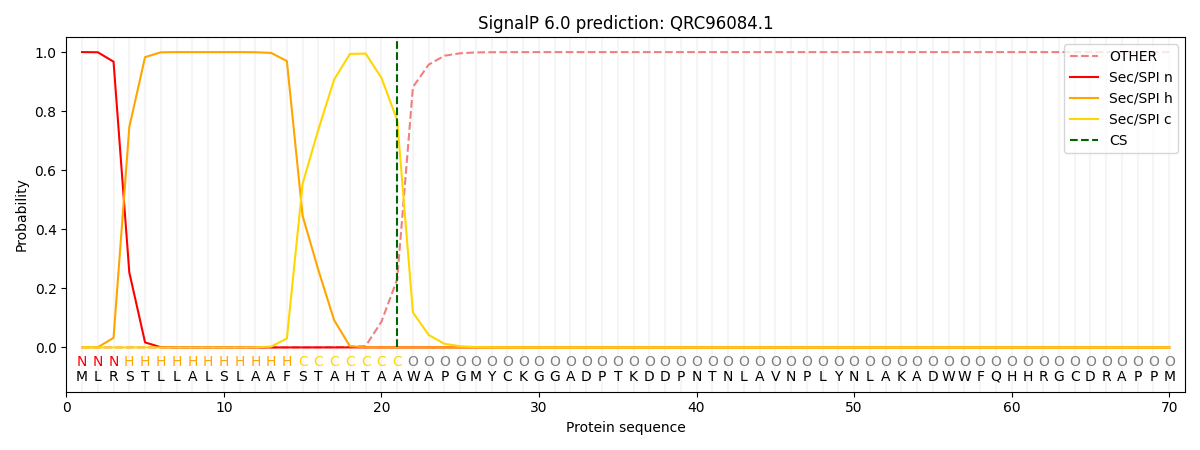
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000267
0.999727
CS pos: 21-22. Pr: 0.7695
There is no transmembrane helices in QRC96084.1.