You are browsing environment: FUNGIDB
CAZyme Information: QRC93519.1
You are here: Home > Sequence: QRC93519.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC93519.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | Carbohydrate-binding module family 18 protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2EV19] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 211316 | ChtBD1_1 | 2.85e-10 | 26 | 69 | 1 | 44 | Hevein or type 1 chitin binding domain; filamentous ascomycete subfamily. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 211316 | ChtBD1_1 | 7.43e-07 | 95 | 140 | 1 | 44 | Hevein or type 1 chitin binding domain; filamentous ascomycete subfamily. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
| 395135 | Chitin_bind_1 | 9.77e-05 | 95 | 133 | 1 | 35 | Chitin recognition protein. |
| 395135 | Chitin_bind_1 | 1.45e-04 | 32 | 61 | 8 | 34 | Chitin recognition protein. |
| 211311 | ChtBD1 | 3.13e-04 | 95 | 135 | 1 | 37 | Hevein or type 1 chitin binding domain. Hevein or type 1 chitin binding domain (ChtBD1), a lectin domain found in proteins from plants and fungi that bind N-acetylglucosamine, plant endochitinases, wound-induced proteins such as hevein, a major IgE-binding allergen in natural rubber latex, and the alpha subunit of Kluyveromyces lactis killer toxin. This domain is involved in the recognition and/or binding of chitin subunits; it typically occurs N-terminal to glycosyl hydrolase domains in chitinases, together with other carbohydrate-binding domains, or by itself in tandem-repeat arrangements. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 615 | 1 | 615 | |
| 1.72e-51 | 9 | 153 | 9 | 166 | |
| 1.77e-48 | 10 | 159 | 9 | 171 | |
| 1.23e-33 | 9 | 150 | 12 | 144 | |
| 2.25e-33 | 9 | 150 | 12 | 144 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.04e-06 | 41 | 61 | 17 | 37 | Crystal structure of the chitin-binding module (CBM18) of a chitinase-like protein from Hevea brasiliensis [Hevea brasiliensis subsp. brasiliensis],4MPI_B Crystal structure of the chitin-binding module (CBM18) of a chitinase-like protein from Hevea brasiliensis [Hevea brasiliensis subsp. brasiliensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.63e-09 | 22 | 68 | 254 | 297 | Chitin deacetylase OS=Pestalotiopsis sp. OX=36460 GN=CDA PE=1 SV=1 |
|
| 9.23e-06 | 25 | 61 | 1 | 35 | Antimicrobial peptide 1 OS=Fagopyrum esculentum OX=3617 PE=1 SV=1 |
|
| 9.23e-06 | 25 | 61 | 1 | 35 | Antimicrobial peptide 2 OS=Fagopyrum esculentum OX=3617 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
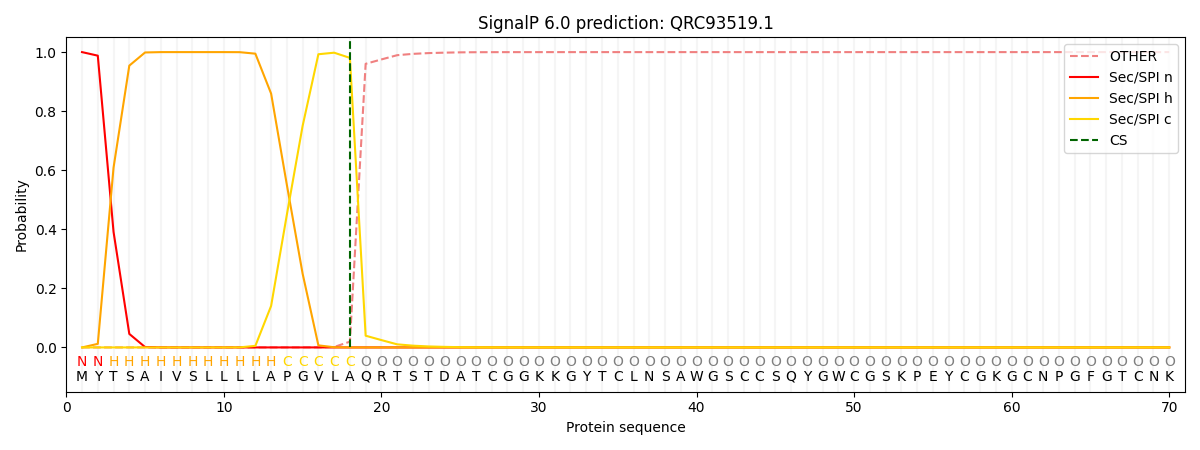
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000266 | 0.999706 | CS pos: 18-19. Pr: 0.9804 |
