You are browsing environment: FUNGIDB
CAZyme Information: QRC91053.1
You are here: Home > Sequence: QRC91053.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC91053.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Glycoside hydrolase family 93 protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2ES79] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2285306; End:2286433 Strand: + | |||||||||||
Full Sequence Download help
| MLLSTLVMSL ISVVSALPQQ QQVQPTFSQK VIFTPPSDYT DPRVLYARSA QLADGTLLAT | 60 |
| WENYSPEPPK VWFPIFQSKD GGNTWSELSR VQDTQQNWGL RYQPFLYVLE NDFPGYAKGT | 120 |
| VLLAGSSIPT DLSQTQIELY ASKDSGATWE FVSHLAAGGE ARPNNGLTPV WEPFLMEYKG | 180 |
| TLIHYYSDQR DNATHGQKMV HQTSSDLKTW GPVIDDVAYP TYTDRPGMPT VALLPNGKYI | 240 |
| MSYEYGGGPA IPSSYQFPVY YKIVDDPEQF GPATGISLKA TDGTVPSGSP YVVWSSVGGA | 300 |
| NGTIIVSAHS GGDIFINKGL GEGPWVKVAT PERSHYTRHL RVLADPTKLL IMGGGQLPPS | 360 |
| TTNKVQFSVM DISNL | 375 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 42 | 352 | 9.4e-111 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 271234 | Sialidase_non-viral | 1.73e-04 | 51 | 250 | 152 | 329 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 271234 | Sialidase_non-viral | 0.005 | 50 | 149 | 20 | 125 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 271234 | Sialidase_non-viral | 0.009 | 50 | 161 | 205 | 305 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 350098 | GH43-like | 0.010 | 135 | 243 | 46 | 150 | Glycosyl hydrolase family 43. This glycosyl hydrolase family 43 (GH43)-like subfamily includes uncharacterized enzymes similar to those with beta-1,4-xylosidase (xylan 1,4-beta-xylosidase; EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanase and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRC91053.1|GH93 | 1.91e-282 | 1 | 375 | 1 | 375 |
| CBX91309.1|GH93 | 9.31e-208 | 3 | 373 | 3 | 375 |
| CAE7032166.1|GH93 | 3.59e-206 | 1 | 373 | 1 | 379 |
| AEO67494.1|GH93|3.2.1.- | 2.83e-205 | 4 | 375 | 4 | 376 |
| AEO55492.1|GH93 | 1.37e-193 | 4 | 375 | 4 | 378 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YDT_A | 3.86e-151 | 26 | 375 | 10 | 364 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
| 2W5N_A | 5.48e-151 | 26 | 375 | 10 | 364 | Chain A, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
| 3A71_A | 2.07e-150 | 32 | 375 | 11 | 354 | High resolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
| 2YDP_A | 3.15e-150 | 26 | 375 | 10 | 364 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
| 2W5O_A | 8.44e-148 | 26 | 375 | 10 | 364 | Chain A, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
SignalP and Lipop Annotations help
This protein is predicted as SP
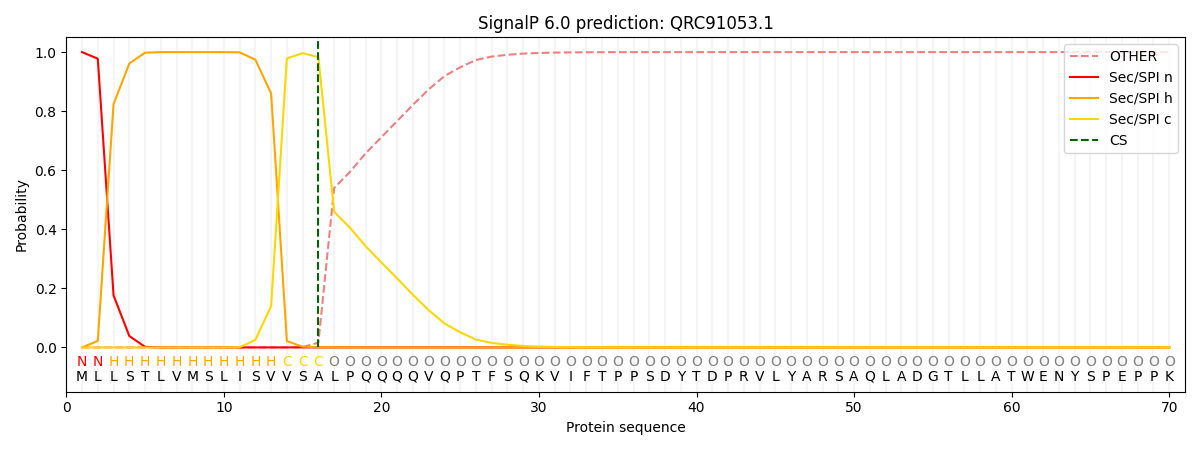
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000268 | 0.999705 | CS pos: 16-17. Pr: 0.9831 |
