You are browsing environment: FUNGIDB
CAZyme Information: QRC90791.1
You are here: Home > Sequence: QRC90791.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC90791.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | FAD-binding PCMH-type domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2EPV3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1747584; End:1749335 Strand: + | |||||||||||
Full Sequence Download help
| MLSSILLTIF CAFLSSTGAA NVADWNSLNK TLKGQLKGAT PLASPCFSQV NGITVTSQAD | 60 |
| KCAGIQAQYT NPRFRVDQFA AYEETQSEDC ATAIGQQCLL DSSDPSSIKA YANVSCNQGS | 120 |
| VPSYYIQVKS AEEVKKAFAF AARTQTAISI KNSGHDYNGR SSGAGSLSLW TRKLQELKYE | 180 |
| PSFVPQQCGR QAGVAAITTG AGINFDQVYT FAHEKGVTYL GGSGPTVGAS GGWVMTGGHG | 240 |
| VLSRVYGLGV DRVLEFEVVT TDGVTRIANA CQNQDLFWAL RGGGGGTFGV ILSTTTRVEP | 300 |
| KLSIAVAFIA LPANTSQQVL AEWTSLAINS TIKWAADGWG GFQGSGITLL GTPLLSLAQA | 360 |
| ESSMAELAQF AKRNGGSVAV ELLSDFYDMY TKYYITNVQA GGSALFSHNW MIPSRAYANA | 420 |
| QGRKQLQDHM DWMSSVGLNP GFLATTPYVY SGKGSTKAKA YAYGPHSSTS TTQAWRSSAA | 480 |
| LLTHSVGWAY NASMSDKKKV ARLLTEASDR ASRLWPDGAS YANEAHPWVK DWKSAFWGGN | 540 |
| YGKLKQIKEK YDSRGLLGCW HCVGSETGGT ADTVGGRCLG KLI | 583 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 122 | 557 | 1.6e-52 | 0.9541484716157205 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 3.59e-21 | 122 | 263 | 1 | 133 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 1.13e-16 | 102 | 557 | 12 | 450 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 273751 | FAD_lactone_ox | 7.27e-07 | 119 | 325 | 12 | 206 | sugar 1,4-lactone oxidases. This model represents a family of at least two different sugar 1,4 lactone oxidases, both involved in synthesizing ascorbic acid or a derivative. These include L-gulonolactone oxidase (EC 1.1.3.8) from rat and D-arabinono-1,4-lactone oxidase (EC 1.1.3.37) from Saccharomyces cerevisiae. Members are proposed to have the cofactor FAD covalently bound at a site specified by Prosite motif PS00862; OX2_COVAL_FAD; 1. |
| 369658 | BBE | 4.04e-06 | 520 | 556 | 1 | 37 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKD57322.1|GH43_11 | 1.39e-39 | 21 | 566 | 511 | 1052 |
| QKD57322.1|CBM91|GH43_11 | 1.39e-39 | 21 | 566 | 511 | 1052 |
| QUC19098.1|AA7 | 7.52e-18 | 82 | 301 | 4 | 217 |
| QRW27390.1|AA7 | 2.55e-15 | 122 | 346 | 66 | 284 |
| AHG26149.1|AA7 | 1.02e-13 | 96 | 333 | 7 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6F72_A | 3.19e-56 | 24 | 566 | 42 | 561 | Crystal structure of VAO-type flavoprotein MtVAO615 at pH 7.5 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_A Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_B Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
| 6F74_A | 4.70e-40 | 25 | 566 | 45 | 585 | Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_B Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_C Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_D Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
| 6FYE_A | 1.01e-15 | 122 | 323 | 45 | 233 | The crystal structure of EncM H138T mutant [Streptomyces maritimus],6FYE_B The crystal structure of EncM H138T mutant [Streptomyces maritimus] |
| 6FYB_A | 1.34e-15 | 122 | 323 | 45 | 233 | The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_B The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_C The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_D The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYC_A The crystal structure of EncM L144M mutant complex with dioxygen under 15 bars O2 pressure [Streptomyces maritimus],6FYC_B The crystal structure of EncM L144M mutant complex with dioxygen under 15 bars O2 pressure [Streptomyces maritimus] |
| 4XLO_A | 1.78e-15 | 122 | 323 | 45 | 233 | Crystal Structure of EncM (crystallized with 4 mM NADPH) [Streptomyces maritimus],4XLO_B Crystal Structure of EncM (crystallized with 4 mM NADPH) [Streptomyces maritimus],4XLO_C Crystal Structure of EncM (crystallized with 4 mM NADPH) [Streptomyces maritimus],4XLO_D Crystal Structure of EncM (crystallized with 4 mM NADPH) [Streptomyces maritimus],6FOQ_A The crystal structure of EncM complexed with dioxygen under 15 bar of oxygen pressure. [Streptomyces maritimus],6FOQ_B The crystal structure of EncM complexed with dioxygen under 15 bar of oxygen pressure. [Streptomyces maritimus],6FOQ_C The crystal structure of EncM complexed with dioxygen under 15 bar of oxygen pressure. [Streptomyces maritimus],6FOQ_D The crystal structure of EncM complexed with dioxygen under 15 bar of oxygen pressure. [Streptomyces maritimus],6FOW_A The crystal structure of EncM complexed with dioxygen under 10 bar of oxygen pressure. [Streptomyces maritimus],6FOW_B The crystal structure of EncM complexed with dioxygen under 10 bar of oxygen pressure. [Streptomyces maritimus],6FOW_C The crystal structure of EncM complexed with dioxygen under 10 bar of oxygen pressure. [Streptomyces maritimus],6FOW_D The crystal structure of EncM complexed with dioxygen under 10 bar of oxygen pressure. [Streptomyces maritimus],6FP3_A The crystal structure of EncM complexed with dioxygen under 5 bar of oxygen pressure. [Streptomyces maritimus],6FP3_B The crystal structure of EncM complexed with dioxygen under 5 bar of oxygen pressure. [Streptomyces maritimus],6FP3_C The crystal structure of EncM complexed with dioxygen under 5 bar of oxygen pressure. [Streptomyces maritimus],6FP3_D The crystal structure of EncM complexed with dioxygen under 5 bar of oxygen pressure. [Streptomyces maritimus],6FY8_A The crystal structure of EncM bromide soak [Streptomyces maritimus],6FY9_A The crystal structure of EncM complex with xenon under 15 bars Xe pressure [Streptomyces maritimus],6FYA_A The crystal structure of EncM under anaerobic conditions [Streptomyces maritimus],6FYA_B The crystal structure of EncM under anaerobic conditions [Streptomyces maritimus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q0V6Q5|PHMC_PHANO | 2.86e-301 | 1 | 452 | 1 | 423 | FAD-linked oxidoreductase phmC OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=phmC PE=3 SV=2 |
| sp|A0A7L8UYL4|FFSJ_ASPFV | 1.45e-235 | 19 | 583 | 20 | 614 | FAD-linked oxidoreductase ffsJ OS=Aspergillus flavipes OX=41900 GN=ffsJ PE=3 SV=1 |
| sp|G4MWA7|OXR2B_MAGO7 | 8.80e-128 | 10 | 582 | 14 | 580 | FAD-linked oxidoreductase OXR2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=OXR2 PE=2 SV=1 |
| sp|P0CU32|CHEF_CHAGB | 7.65e-122 | 18 | 582 | 62 | 612 | FAD-linked oxidoreductase cheF OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=cheF PE=2 SV=1 |
| sp|D4B1Z7|A2478_ARTBC | 2.71e-65 | 25 | 566 | 42 | 560 | Uncharacterized FAD-linked oxidoreductase ARB_02478 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02478 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
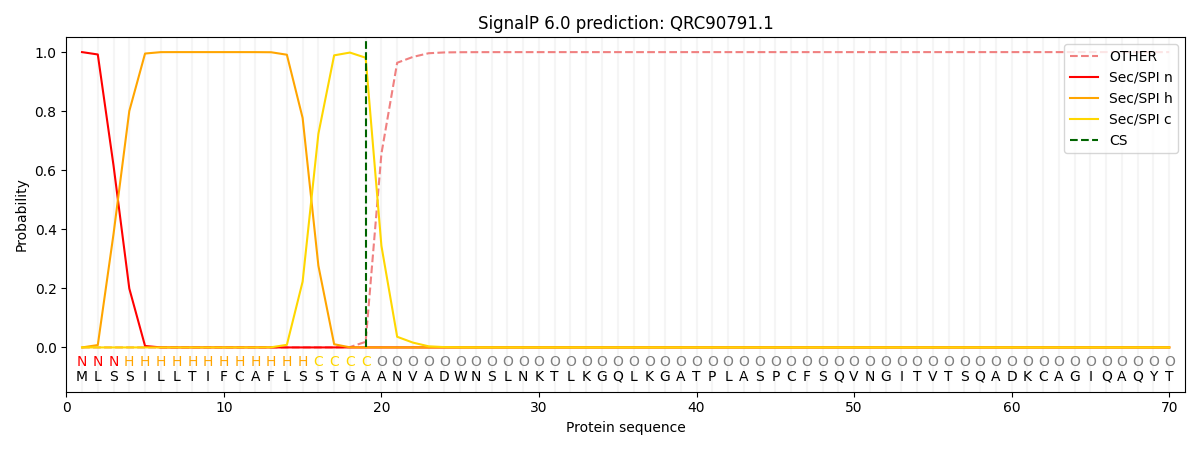
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000210 | 0.999749 | CS pos: 19-20. Pr: 0.9810 |
