You are browsing environment: FUNGIDB
QRC90492.1
Basic Information
help
Species
Parastagonospora nodorum
Lineage
Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum
CAZyme ID
QRC90492.1
CAZy Family
AA2
CAZyme Description
DUF5703_N domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A7U2HWA3]
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
778
CP069023|CGC12 89367.22
5.2482
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_PnodorumSN15
17580
321614
132
17448
Gene Location
No EC number prediction in QRC90492.1.
Family
Start
End
Evalue
family coverage
GH139
10
775
1.8e-246
0.9781209781209781
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
408731
DUF5703_N
8.42e-140
30
312
2
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
778
1
778
0.0
10
778
10
778
0.0
10
778
10
778
0.0
10
778
10
778
0.0
19
778
19
778
QRC90492.1 has no PDB hit.
QRC90492.1 has no Swissprot hit.
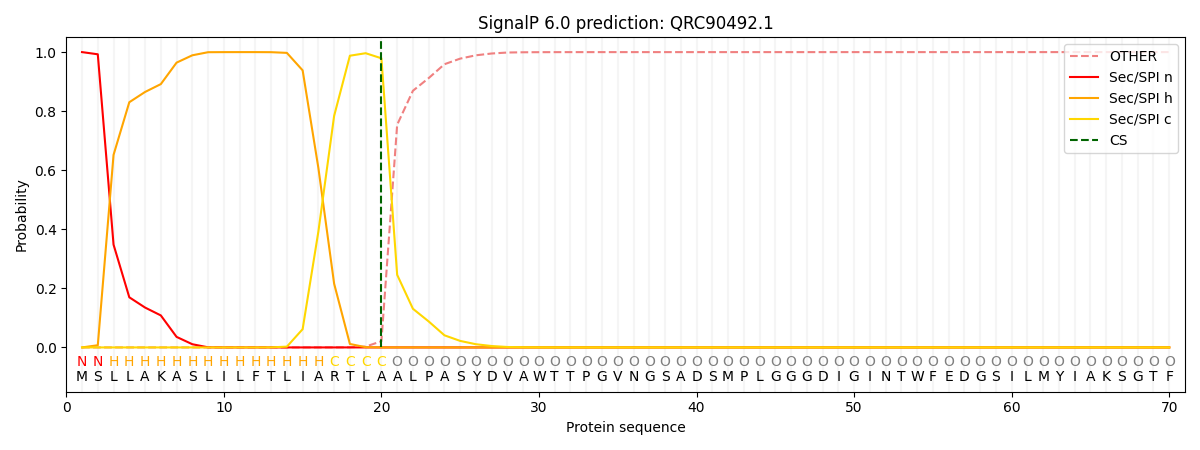
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000235
0.999741
CS pos: 20-21. Pr: 0.9791
There is no transmembrane helices in QRC90492.1.