You are browsing environment: FUNGIDB
CAZyme Information: QRC90244.1
You are here: Home > Sequence: QRC90244.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parastagonospora nodorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Phaeosphaeriaceae; Parastagonospora; Parastagonospora nodorum | |||||||||||
| CAZyme ID | QRC90244.1 | |||||||||||
| CAZy Family | AA11 | |||||||||||
| CAZyme Description | endo-1,4-beta-xylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 44 | 213 | 7.2e-55 | 0.9548022598870056 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 6.06e-80 | 42 | 214 | 3 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.90e-160 | 1 | 221 | 1 | 221 | |
| 2.19e-127 | 1 | 221 | 1 | 223 | |
| 5.14e-126 | 1 | 221 | 1 | 223 | |
| 2.11e-106 | 1 | 219 | 1 | 220 | |
| 5.73e-104 | 1 | 219 | 1 | 219 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.10e-61 | 42 | 211 | 4 | 170 | Chain A, Endo-1,4-beta-xylanase I [Trichoderma reesei] |
|
| 1.84e-44 | 62 | 211 | 35 | 180 | 3D Structure of a thermophilic family GH11 xylanase from Thermobifida fusca [Thermobifida fusca] |
|
| 1.79e-42 | 51 | 219 | 48 | 215 | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE [Escherichia coli] |
|
| 1.91e-42 | 49 | 211 | 25 | 186 | Crystal Structure of a Xylanase at 1.56 Angstroem resolution [Fusarium oxysporum],5JRN_A Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution [Fusarium oxysporum f. sp. vasinfectum 25433] |
|
| 1.96e-42 | 32 | 219 | 20 | 207 | Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus],3WP3_B Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.25e-64 | 1 | 211 | 1 | 221 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_74223 PE=1 SV=1 |
|
| 3.25e-64 | 1 | 211 | 1 | 221 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn1 PE=1 SV=1 |
|
| 5.87e-54 | 1 | 219 | 1 | 216 | Endo-1,4-beta-xylanase 2 OS=Rhizopus oryzae OX=64495 GN=xyn2 PE=1 SV=1 |
|
| 9.95e-52 | 33 | 214 | 39 | 221 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 6.93e-51 | 1 | 211 | 1 | 225 | Probable endo-1,4-beta-xylanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xlnA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
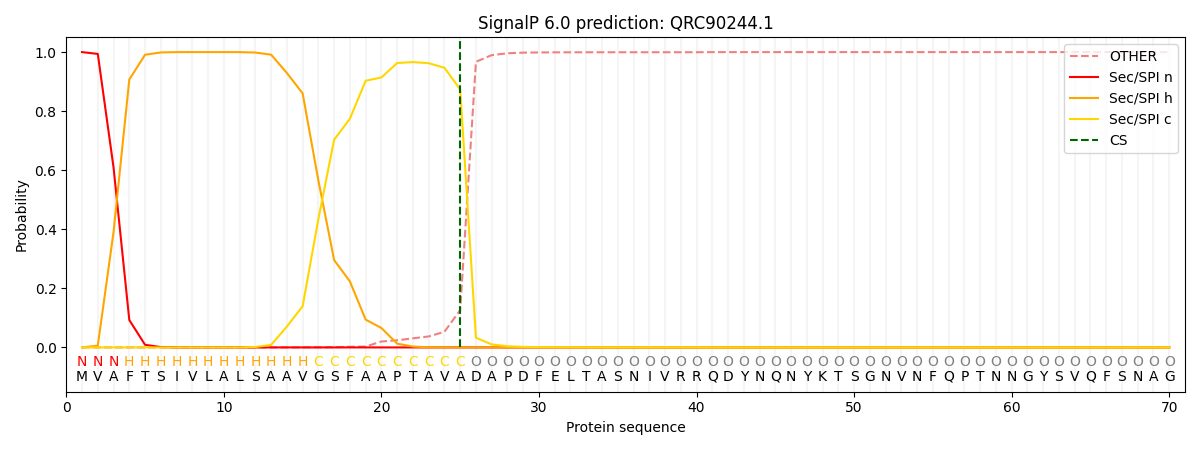
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000299 | 0.999687 | CS pos: 25-26. Pr: 0.8736 |
