You are browsing environment: FUNGIDB
CAZyme Information: QNG14072.1
You are here: Home > Sequence: QNG14072.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] glabrata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] glabrata | |||||||||||
| CAZyme ID | QNG14072.1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH132 | 171 | 469 | 9e-131 | 0.9702970297029703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397780 | SUN | 1.45e-152 | 215 | 456 | 1 | 244 | Beta-glucosidase (SUN family). Members of this family include Nca3, Sun4 and Sim1. This is a family of yeast proteins, involved in a diverse set of functions (DNA replication, aging, mitochondrial biogenesis and cell septation). BGLA from Candida wickerhamii has been characterized as a Beta-glucosidase EC:3.2.1.21. |
| 236090 | PRK07764 | 0.003 | 83 | 170 | 414 | 501 | DNA polymerase III subunits gamma and tau; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.44e-303 | 1 | 469 | 1 | 469 | |
| 5.74e-302 | 1 | 469 | 1 | 469 | |
| 5.74e-302 | 1 | 469 | 1 | 469 | |
| 5.74e-302 | 1 | 469 | 1 | 469 | |
| 5.74e-302 | 1 | 469 | 1 | 469 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.56e-166 | 198 | 469 | 204 | 475 | Probable secreted beta-glucosidase SIM1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SIM1 PE=1 SV=2 |
|
| 2.18e-162 | 197 | 469 | 147 | 419 | Probable secreted beta-glucosidase SUN4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SUN4 PE=1 SV=1 |
|
| 5.07e-135 | 199 | 469 | 147 | 417 | Secreted beta-glucosidase SUN41 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=SUN41 PE=1 SV=2 |
|
| 1.18e-124 | 213 | 469 | 106 | 362 | Probable secreted beta-glucosidase UTH1 OS=Saccharomyces cerevisiae (strain RM11-1a) OX=285006 GN=UTH1 PE=3 SV=1 |
|
| 1.18e-124 | 213 | 469 | 106 | 362 | Probable secreted beta-glucosidase UTH1 OS=Saccharomyces cerevisiae (strain YJM789) OX=307796 GN=UTH1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
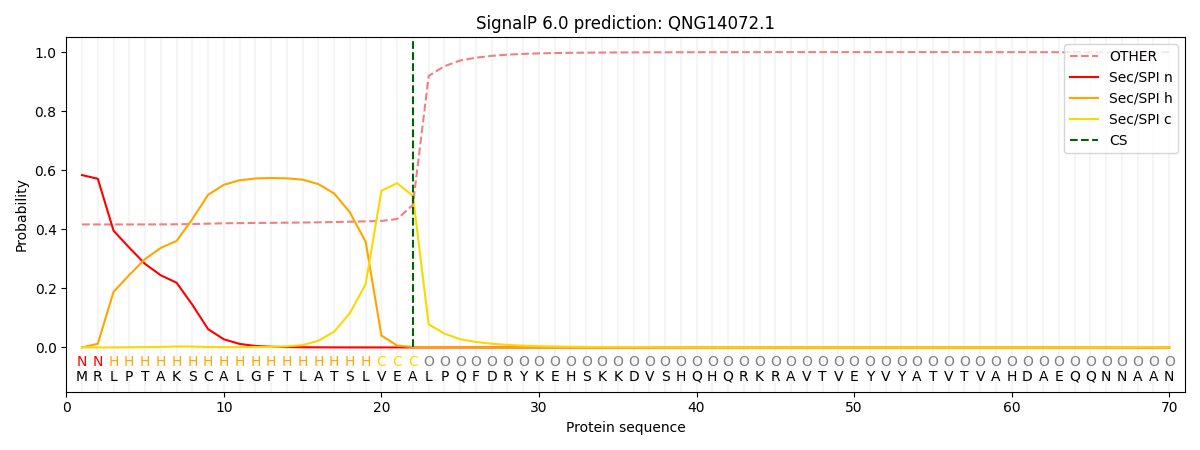
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.427553 | 0.572428 | CS pos: 22-23. Pr: 0.5130 |
