You are browsing environment: FUNGIDB
CAZyme Information: QNG13048.1
Basic Information
help
| Species |
[Candida] glabrata
|
| Lineage |
Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] glabrata
|
| CAZyme ID |
QNG13048.1
|
| CAZy Family |
GT91 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 381 |
CP048233|CGC3 |
44572.16 |
5.3014 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_CglabrataBG2 |
5481 |
N/A |
261 |
5220
|
|
| Gene Location |
No EC number prediction in QNG13048.1.
QNG13048.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.06e-282 |
1 |
381 |
1 |
381 |
| 8.78e-282 |
1 |
381 |
1 |
381 |
| 8.78e-282 |
1 |
381 |
1 |
381 |
| 8.78e-282 |
1 |
381 |
1 |
381 |
| 8.78e-282 |
1 |
381 |
1 |
381 |
QNG13048.1 has no PDB hit.
QNG13048.1 has no Swissprot hit.
This protein is predicted as SP
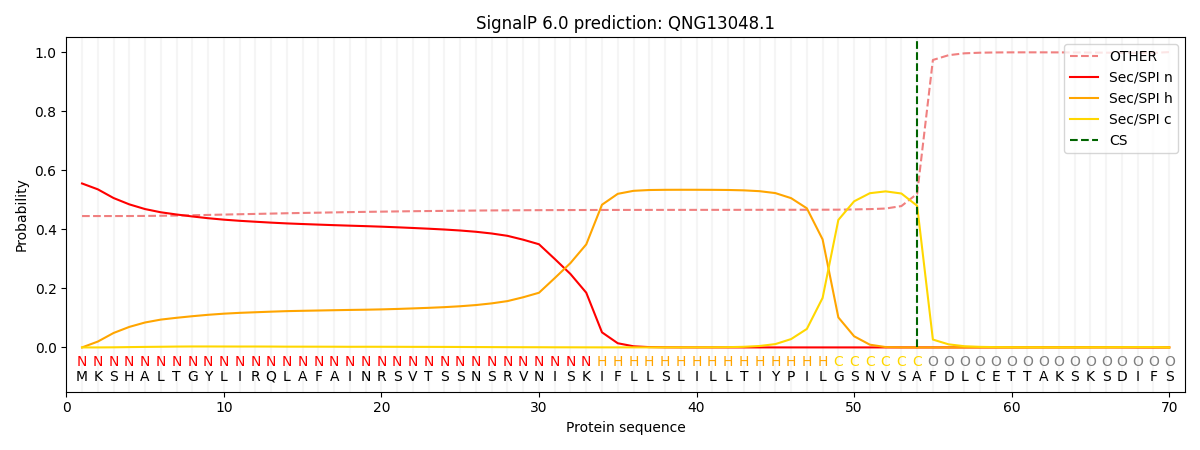
| Other |
SP_Sec_SPI |
CS Position |
| 0.463075 |
0.536918 |
CS pos: 54-55. Pr: 0.4797 |
There is no transmembrane helices in QNG13048.1.

