You are browsing environment: FUNGIDB
CAZyme Information: QHS73923.1
You are here: Home > Sequence: QHS73923.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
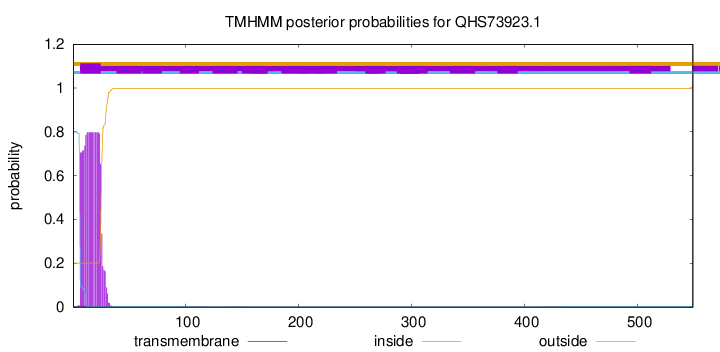
TMHMM annotations
Basic Information help
| Species | Saccharomyces paradoxus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Saccharomycetaceae; Saccharomyces; Saccharomyces paradoxus | |||||||||||
| CAZyme ID | QHS73923.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Sga1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.3:11 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH15 | 111 | 535 | 2.2e-79 | 0.9307479224376731 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225922 | SGA1 | 3.40e-82 | 1 | 548 | 71 | 610 | Glucoamylase (glucan-1,4-alpha-glucosidase), GH15 family [Carbohydrate transport and metabolism]. |
| 395586 | Glyco_hydro_15 | 2.38e-75 | 99 | 535 | 10 | 412 | Glycosyl hydrolases family 15. In higher organisms this family is represented by phosphorylase kinase subunits. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 549 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.59e-98 | 77 | 545 | 14 | 488 | Chain A, GLUCOAMYLASE [Saccharomycopsis fibuligera],2F6D_A Chain A, Glucoamylase GLU1 [Saccharomycopsis fibuligera],2FBA_A Chain A, Glucoamylase GLU1 [Saccharomycopsis fibuligera] |
|
| 1.21e-62 | 79 | 546 | 1 | 433 | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol [Aspergillus niger] |
|
| 1.75e-61 | 79 | 546 | 1 | 432 | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution [Aspergillus awamori],1DOG_A REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION [Aspergillus awamori],1GLM_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori],3GLY_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori] |
|
| 1.79e-61 | 79 | 546 | 1 | 432 | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE [Aspergillus awamori] |
|
| 1.83e-61 | 79 | 546 | 1 | 432 | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE [Aspergillus awamori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 549 | 1 | 549 | Glucoamylase, intracellular sporulation-specific OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SGA1 PE=3 SV=2 |
|
| 4.06e-312 | 29 | 503 | 286 | 760 | Glucoamylase S1 OS=Saccharomyces cerevisiae OX=4932 GN=STA1 PE=3 SV=2 |
|
| 1.39e-310 | 29 | 503 | 287 | 761 | Glucoamylase S2 OS=Saccharomyces cerevisiae OX=4932 GN=STA2 PE=3 SV=1 |
|
| 4.00e-97 | 77 | 545 | 41 | 515 | Glucoamylase GLU1 OS=Saccharomycopsis fibuligera OX=4944 GN=GLU1 PE=1 SV=1 |
|
| 2.42e-95 | 77 | 545 | 41 | 515 | Glucoamylase GLA1 OS=Saccharomycopsis fibuligera OX=4944 GN=GLA1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.038714 | 0.961241 | CS pos: 24-25. Pr: 0.9017 |