You are browsing environment: FUNGIDB
QHS66463.1
Basic Information
help
Species
[Candida] glabrata
Lineage
Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] glabrata
CAZyme ID
QHS66463.1
CAZy Family
GH81
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
1046
CP048125|CGC4 121849.08
5.9478
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CglabrataCBS1382020
5508
N/A
250
5258
Gene Location
No EC number prediction in QHS66463.1.
Family
Start
End
Evalue
family coverage
GH63
458
924
2.2e-18
0.3736842105263158
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
397353
Glyco_hydro_63
1.71e-13
465
924
87
488
Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain.
more
236653
PRK10137
1.08e-04
470
573
350
468
alpha-glucosidase; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
1046
1
1046
0.0
1
1046
1
1046
0.0
1
1046
1
1046
0.0
1
1046
1
1046
0.0
1
1046
1
1046
QHS66463.1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
0.0
1
1034
1
1074
Uncharacterized protein YMR196W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YMR196W PE=1 SV=1
more
1.64e-07
337
930
265
806
Probable mannosyl-oligosaccharide glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC6G10.09 PE=3 SV=1
more
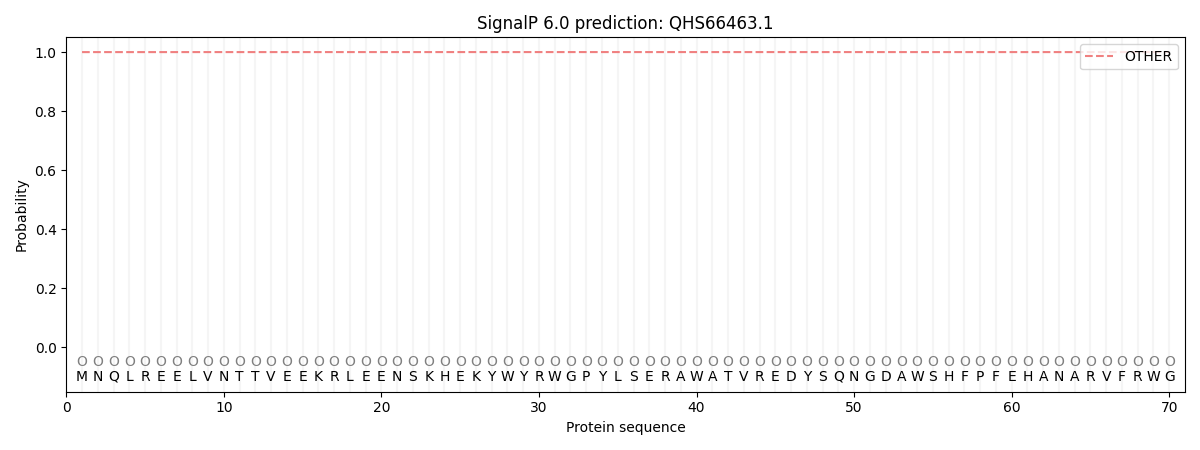
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000057
0.000005
There is no transmembrane helices in QHS66463.1.