You are browsing environment: FUNGIDB
CAZyme Information: PYU1_T007265-p1
Basic Information
help
| Species |
Globisporangium ultimum
|
| Lineage |
Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum
|
| CAZyme ID |
PYU1_T007265-p1
|
| CAZy Family |
GH7 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_GultimumDAOMBR144 |
15430 |
431595 |
140 |
15290
|
|
| Gene Location |
No EC number prediction in PYU1_T007265-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA17 |
12 |
181 |
8.1e-50 |
0.6693877551020408 |
PYU1_T007265-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.09e-46 |
8 |
175 |
9 |
183 |
| 9.97e-46 |
11 |
175 |
6 |
182 |
| 4.90e-36 |
10 |
181 |
9 |
183 |
| 4.90e-36 |
10 |
181 |
9 |
183 |
| 7.33e-36 |
11 |
181 |
10 |
183 |
PYU1_T007265-p1 has no PDB hit.
PYU1_T007265-p1 has no Swissprot hit.
This protein is predicted as SP
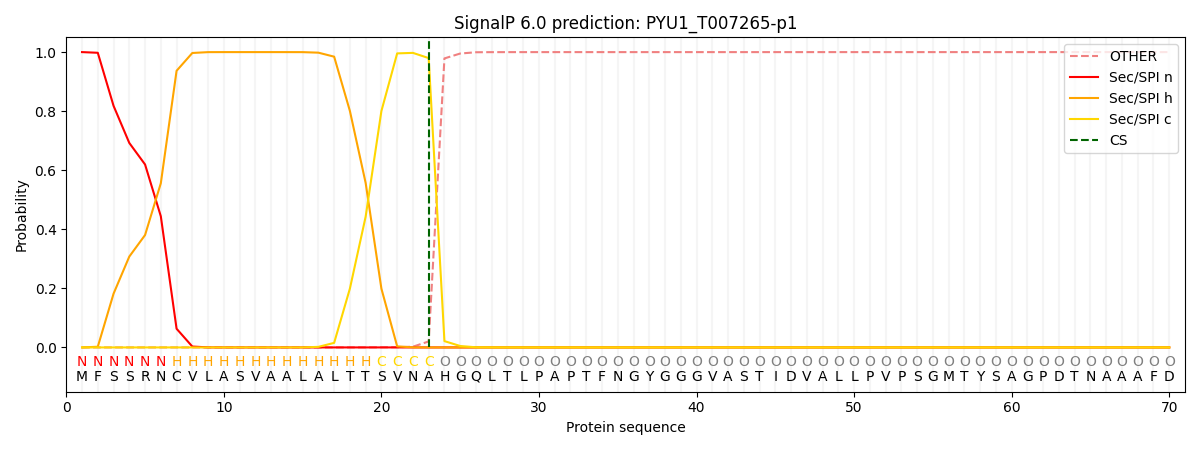
| Other |
SP_Sec_SPI |
CS Position |
| 0.000254 |
0.999723 |
CS pos: 23-24. Pr: 0.9797 |
There is no transmembrane helices in PYU1_T007265-p1.

