You are browsing environment: FUNGIDB
CAZyme Information: PYU1_T006416-p1
You are here: Home > Sequence: PYU1_T006416-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PYU1_T006416-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 550568; End:551386 Strand: - | |||||||||||
Full Sequence Download help
| MNVFATTGAL LLAATPAVLA HGNIVDPPAV WQAGYPQNGF ISEVSSTLWG DMDGATYGYG | 60 |
| PEGAVKYFTK NFPAGSTLKD LILKNQKMYD ASTDPQCGLT IKDEAKRATM PSEVKFSGFS | 120 |
| HPGPCELWCD NNKLAFANDC AATYPTGVVP VDASKCAGAD RFRIFWIGVH SAPWQVYTDC | 180 |
| VYLKGGKSGA TTPATTPKAA GSTPTVTSAA PVANDADDET DAPAATPAPA ADDDEYATPA | 240 |
| PAADDDEYAT DAPAASTPSP TKKKCTGKVR RA | 272 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 10 | 244 | 8.8e-56 | 0.9306122448979591 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402954 | Med3 | 0.009 | 182 | 264 | 124 | 200 | Mediator complex subunit 3 fungal. Mediator is a large complex of up to 33 proteins that is conserved from plants to fungi to humans - the number and representation of individual subunits varying with species. It is arranged into four different sections, a core, a head, a tail and a kinase-activity part, and the number of subunits within each of these is what varies with species. Overall, Mediator regulates the transcriptional activity of RNA polymerase II but it would appear that each of the four different sections has a slightly different function. Mediator subunit Hrs1/Med3 is a physical target for Cyc8-Tup1, a yeast transcriptional co-repressor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EGZ10739.1|AA17 | 1.82e-64 | 7 | 186 | 9 | 193 |
| ETI31514.1|AA17 | 2.22e-63 | 7 | 186 | 9 | 193 |
| ETK71917.1|AA17 | 2.22e-63 | 7 | 186 | 9 | 193 |
| ETI31524.1|AA17 | 4.48e-63 | 7 | 195 | 9 | 202 |
| ETP29504.1|AA17 | 8.63e-63 | 7 | 186 | 9 | 193 |
SignalP and Lipop Annotations help
This protein is predicted as SP
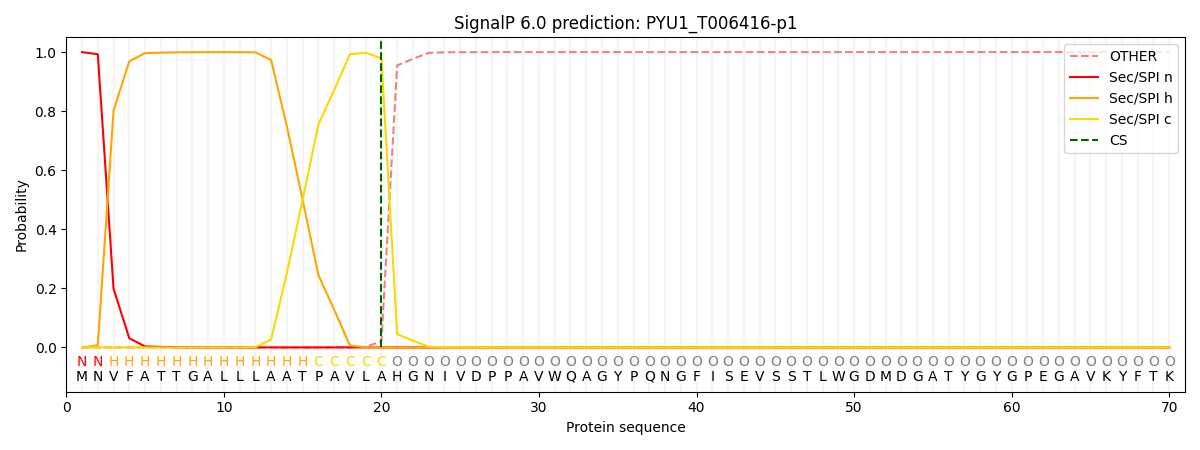
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000221 | 0.999740 | CS pos: 20-21. Pr: 0.9786 |
