You are browsing environment: FUNGIDB
CAZyme Information: PYU1_T005915-p1
You are here: Home > Sequence: PYU1_T005915-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
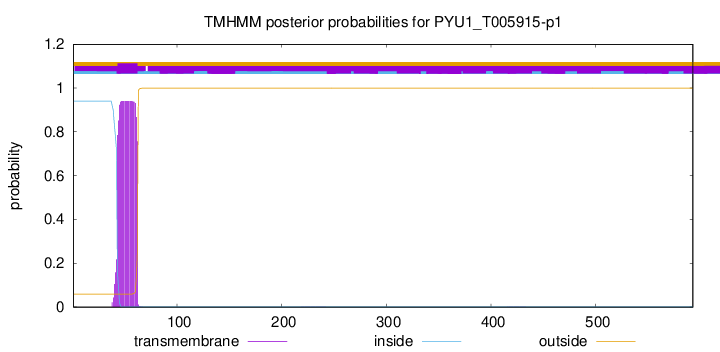
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PYU1_T005915-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:99 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 150 | 585 | 5.2e-134 | 0.9955156950672646 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 1.52e-179 | 150 | 585 | 1 | 452 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 9.54e-172 | 135 | 585 | 64 | 517 | glycoside hydrolase family 47 protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.24e-218 | 43 | 588 | 47 | 608 | |
| 2.67e-153 | 139 | 589 | 70 | 509 | |
| 8.69e-130 | 61 | 586 | 124 | 641 | |
| 2.65e-129 | 121 | 586 | 164 | 636 | |
| 1.37e-128 | 137 | 589 | 3 | 458 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.51e-129 | 144 | 589 | 11 | 459 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 8.49e-129 | 144 | 586 | 6 | 451 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 1.03e-128 | 144 | 586 | 11 | 456 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 2.64e-128 | 144 | 589 | 6 | 454 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
|
| 4.59e-128 | 132 | 589 | 77 | 537 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.63e-128 | 129 | 589 | 193 | 656 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Rattus norvegicus OX=10116 GN=Man1b1 PE=2 SV=2 |
|
| 1.88e-127 | 128 | 589 | 193 | 657 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Mus musculus OX=10090 GN=Man1b1 PE=1 SV=1 |
|
| 2.59e-125 | 132 | 589 | 238 | 698 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Homo sapiens OX=9606 GN=MAN1B1 PE=1 SV=2 |
|
| 2.94e-109 | 144 | 588 | 129 | 622 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 OS=Arabidopsis thaliana OX=3702 GN=MNS3 PE=1 SV=1 |
|
| 1.39e-95 | 144 | 585 | 39 | 507 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Candida albicans OX=5476 GN=MNS1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
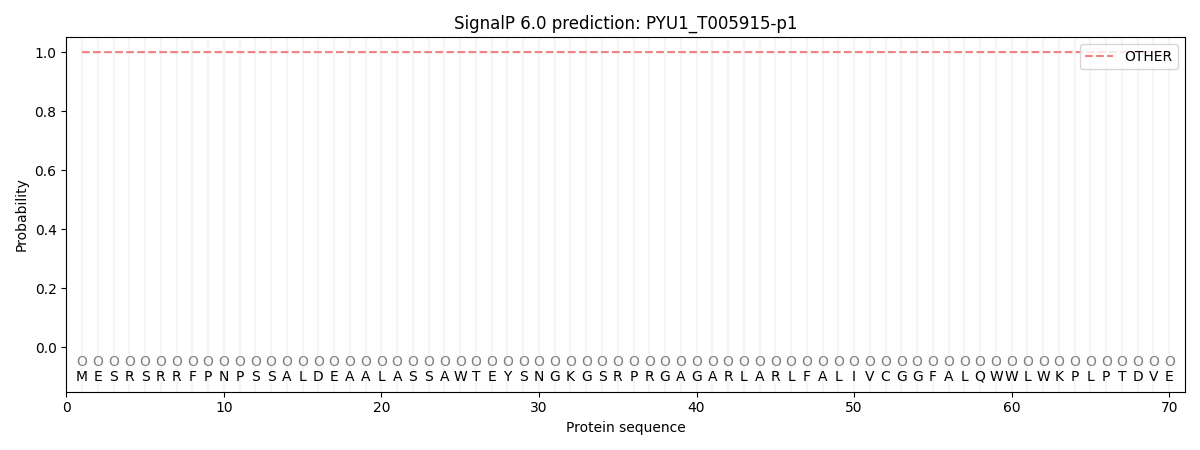
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999946 | 0.000048 |