You are browsing environment: FUNGIDB
CAZyme Information: PYU1_T005551-p1
You are here: Home > Sequence: PYU1_T005551-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PYU1_T005551-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 60256; End:63908 Strand: + | |||||||||||
Full Sequence Download help
| MPKIAMAVCA RTAARRRPFS RRTAAVRKAL TLLLLVVCGW FAGAGSSASE ASYEDMFPTA | 60 |
| TKTLLEIVHP PNDLVLVGSD LRIEILIRNE LAAELVDSKV CISMDPVFIP DDVQLDEGSQ | 120 |
| LHESCFDHST ENYTTFNIDG LVPGLAYGVT VGLISHAKIL GISMRTFEVG SIILPTLGSR | 180 |
| VSIASALETG FEYQKLGNRQ EAASIYSLVL DLFPDHPRAM HLLGFMLCQE DNPHRGYAYI | 240 |
| YRAVQANTSE WSFAISLGMC LTNMRNFTEA IQYYRKALEL HPASYDAALN MGDAFQAMGN | 300 |
| WDDALHEYRK VAVSASGAYQ EPTSRIEDPD KYVADALGRV CETTRVMEGG FACEKCLVEA | 360 |
| ITRFPDDAQL RNDHGNLLLS AGRFESALQE YHSASNLGSL FGMVRALLLV QAGIYKTLTC | 420 |
| SISVADALEI LGETYESLEQ YELILRTLNS EQGYPGTRIR IMKATVLPRI LPGSQQEIDM | 480 |
| YRQRFESEVD ALRDDIETLQ PTDMEPSRIP FSTAVTMNSH NRNNRLLKTK IGTLYFDLLY | 540 |
| TRRLLREKYM LGYAAIPSPD RFIRPSGVPT QRRLRVGFVS RFLFSPTVGL YMSNLIPALD | 600 |
| PLKYQTIAFA IGLSSSMKKR EHAGRIAEEV WALPKDLPTA RNEIRAANLD VLIYPELGMD | 660 |
| KTTYFLSLLR LAPIQAVWWG NTDTSGVQAI DYRLVSEYEH ENAREHYTEK ELYQFNFPEP | 720 |
| RDYNASRTYV RDAVIKRFNL PADFHMYFSL ESILNMHPDY DEAVVEILRR DDKARMFFLG | 780 |
| SSTRNRWRDQ LVARIVVRAG DSVRAFERIY FMNDVDSKQE LLLAGAADTV MASVHLTRPH | 840 |
| ATIQAFTAGV PVVTLPGEFW GTRVAYGFYQ IMEMNNLIAS NMDEYVTLAV RMADDADFRN | 900 |
| EMMEKIKERR SRLHEDSRAV EEWEKFLDFA GAKLYPSGED NDYEVSDSKG DEESNTQDQT | 960 |
| EENELTCNKS DEDENAQTCL REEKRAK | 987 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 352 | 926 | 1.4e-62 | 0.5205673758865248 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 4.78e-27 | 422 | 911 | 105 | 593 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 2.51e-14 | 217 | 311 | 1 | 95 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 8.78e-13 | 190 | 279 | 8 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 223533 | TPR | 1.57e-08 | 193 | 321 | 140 | 272 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
| 404688 | Glyco_transf_41 | 9.85e-08 | 807 | 911 | 425 | 529 | Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UIZ20641.1|GT41 | 8.09e-182 | 47 | 950 | 20 | 923 |
| BAZ61948.1|GT41 | 2.40e-50 | 444 | 930 | 427 | 884 |
| BAZ15802.1|GT41 | 2.40e-50 | 444 | 930 | 427 | 884 |
| BBO73735.1|GT41 | 1.45e-48 | 461 | 935 | 374 | 819 |
| BBO88349.1|GT41 | 6.76e-48 | 460 | 935 | 429 | 877 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JLB_A | 2.89e-07 | 555 | 895 | 187 | 522 | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
| 2VSN_A | 2.89e-07 | 555 | 895 | 187 | 522 | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
| 5A01_A | 3.26e-07 | 736 | 906 | 504 | 668 | O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_B O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster],5A01_C O-GlcNAc transferase from Drososphila melanogaster [Drosophila melanogaster] |
SignalP and Lipop Annotations help
This protein is predicted as SP
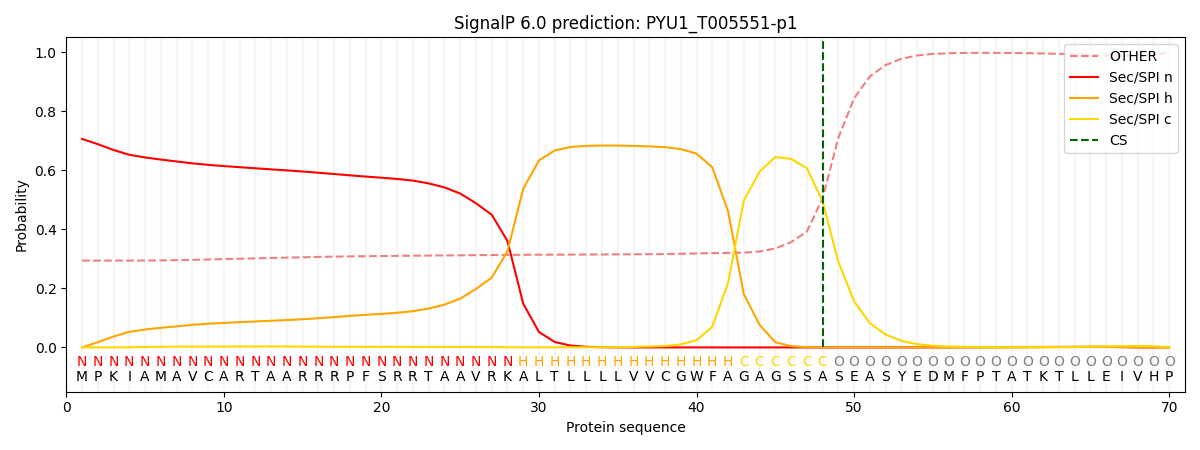
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.319856 | 0.680140 | CS pos: 48-49. Pr: 0.4947 |

