You are browsing environment: FUNGIDB
CAZyme Information: PYU1_T001521-p1
You are here: Home > Sequence: PYU1_T001521-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PYU1_T001521-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA16 | 21 | 188 | 6.1e-22 | 0.9880239520958084 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 3.85e-08 | 21 | 189 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| 411340 | gliding_GltC | 0.001 | 204 | 248 | 34 | 81 | adventurous gliding motility protein GltC. GltC is a soluble periplasmic protein required for a type of gliding motility found in certain social delta-proteobacteria, including the model species Myxococcus xanthus. |
| 410624 | LPMO_AA10 | 0.005 | 21 | 120 | 1 | 113 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| 411416 | MCP_Sipho | 0.006 | 206 | 246 | 77 | 117 | major capsid protein, Siphoviridae type. This protein is a phage major capsid protein, as reported in primary sequence submissions of a large number of Siphoviridae, many of which have hosts in the Mycobacterium and Gordonia genera of bacteria. |
| 227315 | FabG2 | 0.009 | 206 | 245 | 6 | 45 | 3-oxoacyl-ACP reductase domain of yeast-type FAS1 [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.36e-15 | 21 | 188 | 18 | 189 | |
| 2.58e-15 | 6 | 196 | 3 | 193 | |
| 2.58e-15 | 6 | 196 | 3 | 193 | |
| 6.78e-15 | 7 | 188 | 5 | 191 | |
| 6.93e-14 | 1 | 211 | 1 | 206 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
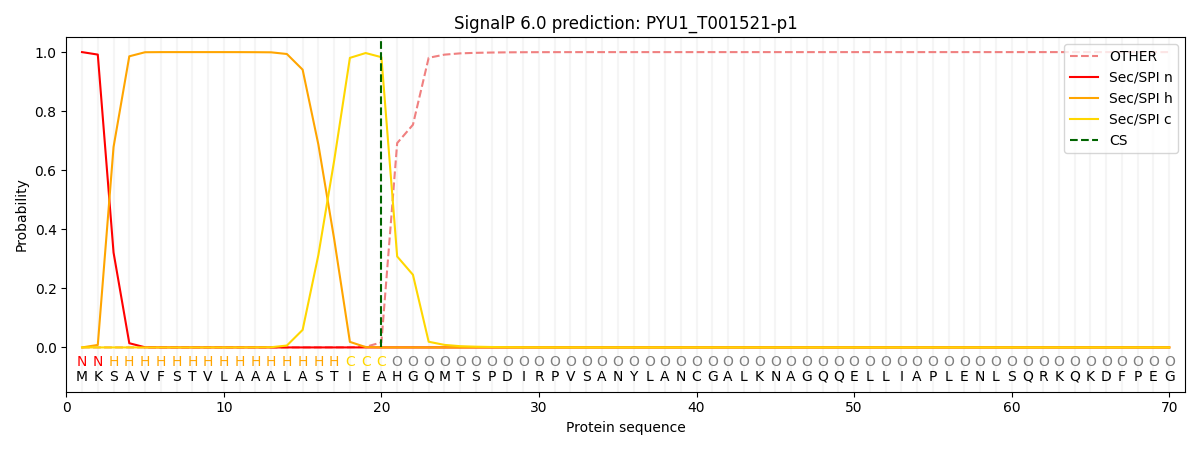
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000227 | 0.999745 | CS pos: 20-21. Pr: 0.9828 |
