You are browsing environment: FUNGIDB
CAZyme Information: PWY91217.1
You are here: Home > Sequence: PWY91217.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus heteromorphus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus heteromorphus | |||||||||||
| CAZyme ID | PWY91217.1 | |||||||||||
| CAZy Family | GT33 | |||||||||||
| CAZyme Description | cellulose-binding GDSL lipase/acylhydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE16 | 13 | 264 | 5.6e-98 | 0.9775280898876404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238882 | fatty_acyltransferase_like | 8.26e-63 | 13 | 264 | 6 | 267 | Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. Might catalyze fatty acid transfer between phosphatidylcholine and sterols. |
| 238875 | SGNH_plant_lipase_like | 1.05e-26 | 67 | 252 | 79 | 299 | SGNH_plant_lipase_like, a plant specific subfamily of the SGNH-family of hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 225780 | COG3240 | 2.22e-10 | 13 | 264 | 36 | 329 | Phospholipase/lecithinase/hemolysin [Lipid transport and metabolism, General function prediction only]. |
| 395531 | Lipase_GDSL | 2.23e-09 | 13 | 262 | 5 | 222 | GDSL-like Lipase/Acylhydrolase. |
| 238883 | Triacylglycerol_lipase_like | 1.27e-06 | 13 | 235 | 8 | 231 | Triacylglycerol lipase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. Members of this subfamily might hydrolyze triacylglycerol into diacylglycerol and fatty acid anions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.32e-149 | 5 | 274 | 22 | 293 | |
| 3.98e-148 | 4 | 274 | 38 | 310 | |
| 8.58e-133 | 4 | 274 | 27 | 298 | |
| 8.58e-133 | 4 | 274 | 27 | 298 | |
| 8.58e-133 | 4 | 274 | 27 | 298 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.71e-62 | 13 | 262 | 38 | 291 | Acetylesterase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=aes1 PE=3 SV=1 |
|
| 1.05e-61 | 13 | 262 | 38 | 291 | Acetylesterase OS=Thermothelomyces thermophilus OX=78579 GN=aes1 PE=1 SV=1 |
|
| 1.89e-07 | 87 | 268 | 161 | 379 | GDSL esterase/lipase EXL2 OS=Arabidopsis thaliana OX=3702 GN=EXL2 PE=2 SV=1 |
|
| 3.54e-07 | 133 | 267 | 236 | 399 | GDSL esterase/lipase At1g20120 OS=Arabidopsis thaliana OX=3702 GN=At1g20120 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
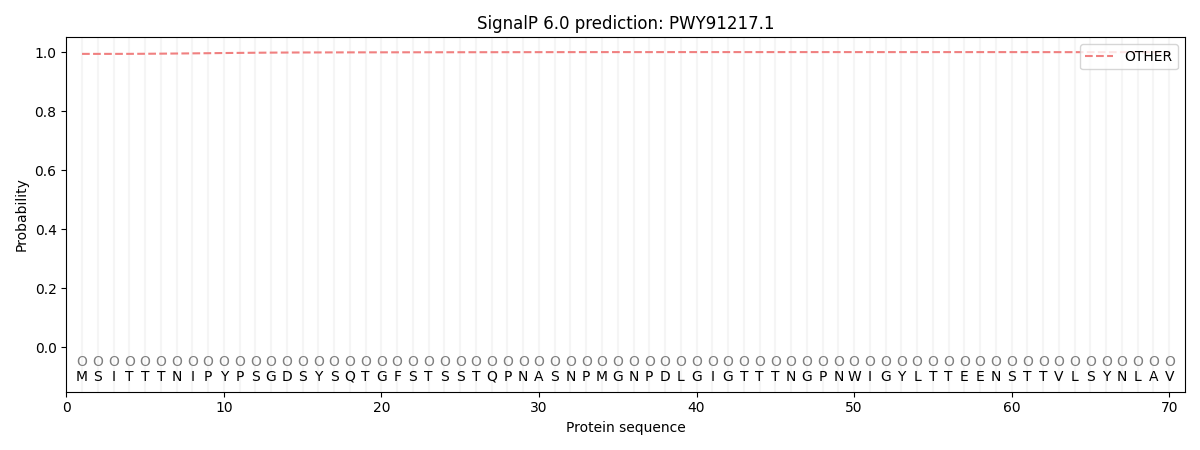
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.994403 | 0.005627 |
