You are browsing environment: FUNGIDB
CAZyme Information: PWY89150.1
You are here: Home > Sequence: PWY89150.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
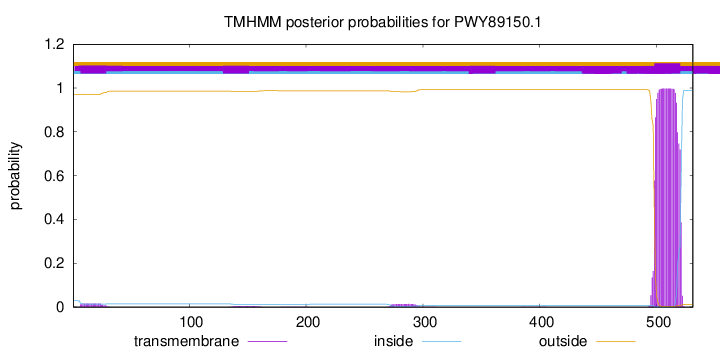
TMHMM annotations
Basic Information help
| Species | Aspergillus heteromorphus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus heteromorphus | |||||||||||
| CAZyme ID | PWY89150.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | putative UDP-glucoronosyl and UDP-glucosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 116 | 432 | 2.8e-26 | 0.7041884816753927 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 2.11e-40 | 10 | 461 | 4 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 7.17e-16 | 7 | 463 | 3 | 399 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 278624 | UDPGT | 1.73e-10 | 351 | 444 | 330 | 421 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 215465 | PLN02863 | 1.03e-09 | 19 | 401 | 22 | 401 | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
| 223071 | egt | 3.85e-08 | 352 | 444 | 355 | 445 | ecdysteroid UDP-glucosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.88e-231 | 2 | 524 | 6 | 530 | |
| 9.03e-231 | 5 | 528 | 5 | 527 | |
| 1.47e-223 | 4 | 524 | 7 | 530 | |
| 1.18e-216 | 4 | 524 | 9 | 511 | |
| 2.33e-213 | 4 | 525 | 8 | 531 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.95e-09 | 352 | 444 | 99 | 189 | Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_B Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_C Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_D Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens] |
|
| 5.44e-08 | 352 | 429 | 78 | 155 | Chain A, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_B Chain B, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_C Chain C, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_D Chain D, UDP-glucuronosyltransferase 2B15 [Homo sapiens] |
|
| 5.44e-06 | 116 | 422 | 136 | 449 | Structure of Phytolacca americana UGT3 with 15-crown-5 [Phytolacca americana],6LZX_B Structure of Phytolacca americana UGT3 with 15-crown-5 [Phytolacca americana],6LZY_A Structure of Phytolacca americana UGT3 with 18-crown-6 [Phytolacca americana],6LZY_B Structure of Phytolacca americana UGT3 with 18-crown-6 [Phytolacca americana],7VEJ_A Chain A, Glycosyltransferase [Phytolacca americana],7VEJ_B Chain B, Glycosyltransferase [Phytolacca americana],7VEK_A Chain A, Glycosyltransferase [Phytolacca americana],7VEK_B Chain B, Glycosyltransferase [Phytolacca americana],7VEL_A Chain A, Glycosyltransferase [Phytolacca americana],7VEL_B Chain B, Glycosyltransferase [Phytolacca americana] |
|
| 8.65e-06 | 350 | 468 | 324 | 437 | Structural Characterization of UDP-glycosyltransferase from Tetranychus Urticae [Tetranychus urticae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.42e-11 | 152 | 444 | 187 | 448 | UDP-glucuronosyltransferase 2B4 OS=Homo sapiens OX=9606 GN=UGT2B4 PE=1 SV=2 |
|
| 1.33e-10 | 152 | 444 | 181 | 442 | UDP-glucuronosyltransferase 2B16 (Fragment) OS=Oryctolagus cuniculus OX=9986 GN=UGT2B16 PE=2 SV=2 |
|
| 1.78e-10 | 110 | 444 | 149 | 449 | UDP-glucuronosyltransferase 2A3 OS=Cavia porcellus OX=10141 GN=UGT2A3 PE=2 SV=1 |
|
| 5.45e-10 | 121 | 444 | 159 | 448 | UDP-glucuronosyltransferase 2B33 OS=Macaca mulatta OX=9544 GN=UGT2B33 PE=1 SV=1 |
|
| 5.45e-10 | 121 | 444 | 159 | 448 | UDP-glucuronosyltransferase 2B23 OS=Macaca fascicularis OX=9541 GN=UGT2B23 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
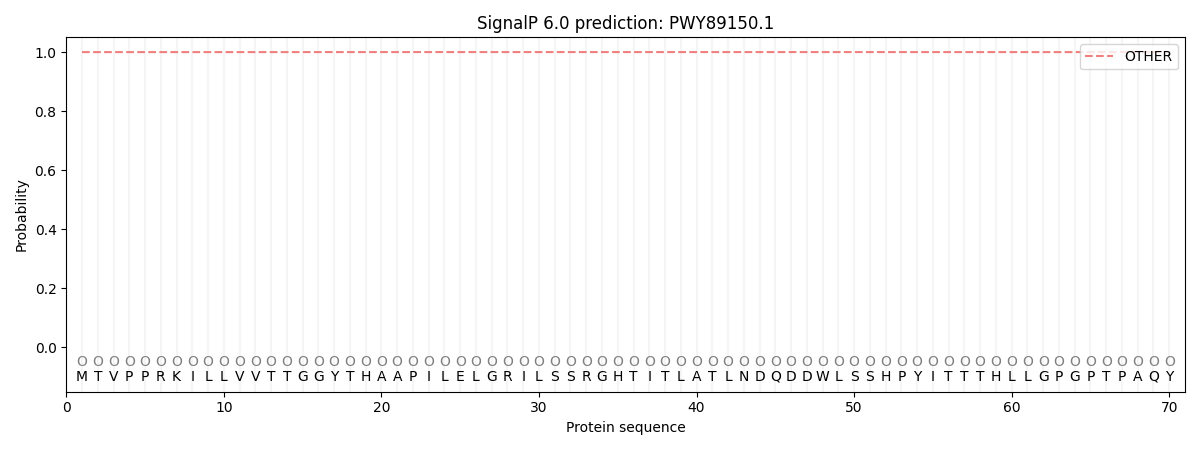
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000002 |