You are browsing environment: FUNGIDB
CAZyme Information: PWY75014.1
You are here: Home > Sequence: PWY75014.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus heteromorphus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus heteromorphus | |||||||||||
| CAZyme ID | PWY75014.1 | |||||||||||
| CAZy Family | GH114 | |||||||||||
| CAZyme Description | Glucan 1,4-alpha-glucosidase [Source:UniProtKB/TrEMBL;Acc:A0A317VKX0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.3:94 | 3.2.1.3:71 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH15 | 43 | 448 | 8.8e-79 | 0.9806094182825484 |
| CBM20 | 533 | 611 | 9.4e-27 | 0.8333333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 293766 | G6S | 0.0 | 654 | 1106 | 1 | 394 | glucosamine (N-acetyl)-6-sulfatase(G6S, GNS) AND sulfatase 1(SULF1). N-acetylglucosamine-6-sulfatase also known as glucosamine (N-acetyl)-6-sulfatase hydrolyzes of the 6-sulfate groups of the N-acetyl-D-glucosamine 6-sulfate units of heparan sulfate and keratan sulfate. Deficient of N-acetylglucosamine-6-sulfatase results in disease of Sanfilippo Syndrome type IIId or Mucopolysaccharidosis III (MPS-III), a rare autosomal recessive lysosomal storage disease. SULF1 encodes an extracellular heparan sulfate endosulfatase, that removes 6-O-sulfate groups from heparan sulfate chains of heparan sulfate proteoglycans (HSPGs). |
| 395586 | Glyco_hydro_15 | 3.11e-144 | 37 | 450 | 2 | 417 | Glycosyl hydrolases family 15. In higher organisms this family is represented by phosphorylase kinase subunits. |
| 293755 | G6S_like | 2.44e-71 | 653 | 1111 | 1 | 418 | unchracterized sulfatase homologous to glucosamine (N-acetyl)-6-sulfatase(G6S, GNS). N-acetylglucosamine-6-sulfatase also known as glucosamine (N-acetyl)-6-sulfatase hydrolyzes of the 6-sulfate groups of the N-acetyl-D-glucosamine 6-sulfate units of heparan sulfate and keratan sulfate. Deficiency of N-acetylglucosamine-6-sulfatase results in the disease of Sanfilippo Syndrome type IIId or Mucopolysaccharidosis III (MPS-III), a rare autosomal recessive lysosomal storage disease. |
| 293763 | ARS_like | 7.28e-49 | 655 | 1109 | 1 | 411 | uncharacterized arylsulfatase subfamily. Sulfatases catalyze the hydrolysis of sulfate esters from wide range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. The biological roles of sulfatase includes the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodeling sulfated glycosaminoglycans in the extracellular space. The sulfatases are essential for human metabolism. At least eight human monogenic diseases are caused by the deficiency of individual sulfatases. |
| 395712 | Sulfatase | 2.52e-48 | 655 | 1004 | 1 | 296 | Sulfatase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 27 | 1191 | 36 | 1236 | |
| 0.0 | 1 | 610 | 1 | 615 | |
| 0.0 | 1 | 610 | 1 | 619 | |
| 0.0 | 1 | 610 | 1 | 615 | |
| 0.0 | 1 | 610 | 1 | 615 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 25 | 610 | 1 | 592 | Structure of the catalytic domain of Aspergillus niger Glucoamylase [Aspergillus niger] |
|
| 3.75e-301 | 25 | 496 | 1 | 472 | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE [Aspergillus awamori] |
|
| 2.91e-300 | 25 | 495 | 1 | 471 | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE [Aspergillus awamori] |
|
| 3.97e-300 | 25 | 493 | 1 | 469 | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution [Aspergillus awamori],1DOG_A REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION [Aspergillus awamori],1GLM_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori],3GLY_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori] |
|
| 1.13e-299 | 25 | 493 | 1 | 470 | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 610 | 1 | 615 | Glucoamylase OS=Aspergillus usamii OX=186680 GN=glaA PE=3 SV=1 |
|
| 0.0 | 1 | 610 | 1 | 616 | Glucoamylase OS=Aspergillus niger OX=5061 GN=GLAA PE=1 SV=1 |
|
| 0.0 | 1 | 610 | 1 | 616 | Glucoamylase OS=Aspergillus awamori OX=105351 GN=GLAA PE=1 SV=1 |
|
| 0.0 | 1 | 610 | 1 | 615 | Glucoamylase I OS=Aspergillus kawachii OX=1069201 GN=gaI PE=1 SV=1 |
|
| 1.36e-284 | 20 | 610 | 23 | 588 | Glucoamylase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=glaA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
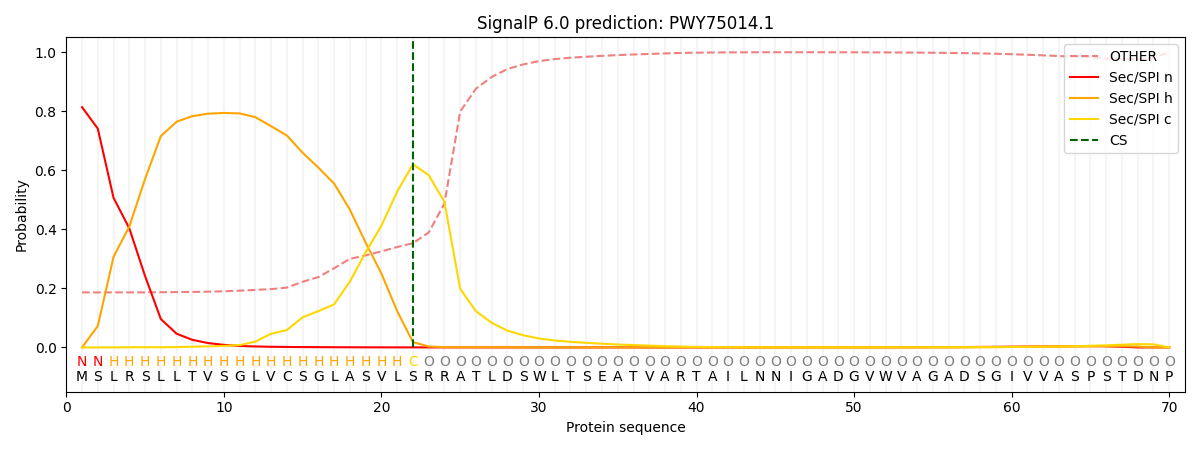
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.204509 | 0.795465 | CS pos: 22-23. Pr: 0.6199 |
