You are browsing environment: FUNGIDB
CAZyme Information: PWY74326.1
Basic Information
help
| Species |
Aspergillus heteromorphus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus heteromorphus
|
| CAZyme ID |
PWY74326.1
|
| CAZy Family |
GH1 |
| CAZyme Description |
glycoside hydrolase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 326 |
|
35954.45 |
3.7661 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AheteromorphusCBS117.55 |
11436 |
1448321 |
306 |
11130
|
|
| Gene Location |
No EC number prediction in PWY74326.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
84 |
271 |
2.1e-19 |
0.5412541254125413 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 226609
|
ManB2 |
5.78e-09 |
122 |
257 |
167 |
300 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| 396639
|
Glyco_hydro_26 |
0.005 |
190 |
258 |
203 |
262 |
Glycosyl hydrolase family 26. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.39e-59 |
34 |
325 |
36 |
316 |
| 1.64e-58 |
34 |
325 |
83 |
363 |
| 1.71e-54 |
9 |
311 |
9 |
301 |
| 2.51e-51 |
3 |
325 |
11 |
317 |
| 2.93e-51 |
34 |
318 |
32 |
307 |
PWY74326.1 has no PDB hit.
PWY74326.1 has no Swissprot hit.
This protein is predicted as SP
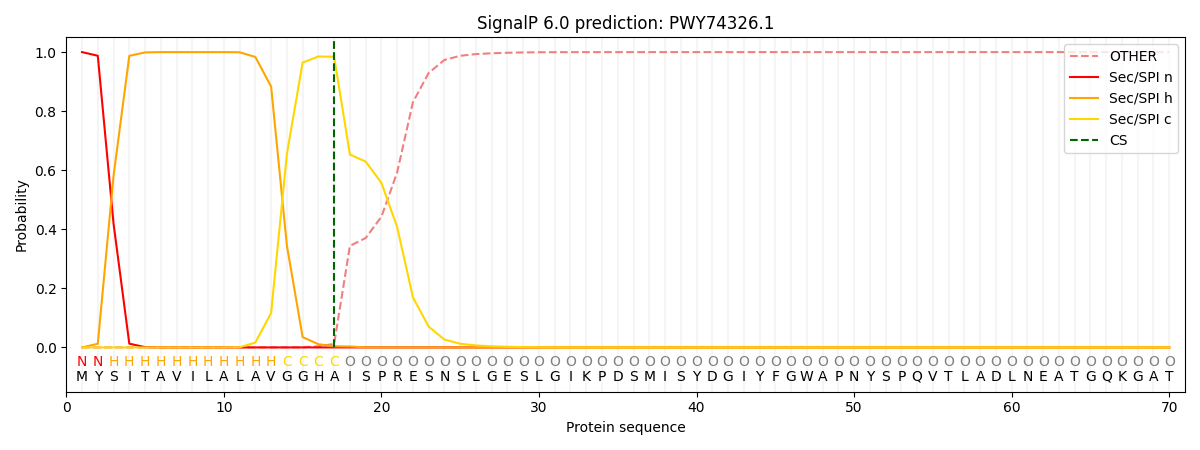
| Other |
SP_Sec_SPI |
CS Position |
| 0.000213 |
0.999758 |
CS pos: 17-18. Pr: 0.9841 |
There is no transmembrane helices in PWY74326.1.

