You are browsing environment: FUNGIDB
CAZyme Information: PWY71349.1
You are here: Home > Sequence: PWY71349.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus eucalypticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus eucalypticola | |||||||||||
| CAZyme ID | PWY71349.1 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 72 | 257 | 4.7e-30 | 0.7870370370370371 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396478 | Glyco_hydro_3_C | 1.05e-41 | 326 | 568 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| 185053 | PRK15098 | 1.25e-23 | 86 | 661 | 120 | 729 | beta-glucosidase BglX. |
| 405066 | Fn3-like | 2.20e-12 | 618 | 675 | 1 | 70 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
| 395747 | Glyco_hydro_3 | 2.87e-10 | 65 | 237 | 69 | 227 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.10e-226 | 8 | 687 | 9 | 746 | |
| 4.83e-224 | 27 | 687 | 32 | 743 | |
| 5.66e-224 | 6 | 688 | 1 | 791 | |
| 5.66e-224 | 6 | 688 | 1 | 791 | |
| 5.24e-222 | 1 | 684 | 1 | 745 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.00e-92 | 31 | 687 | 10 | 712 | Chain A, Beta-d-glucoside Glucohydrolase [Trichoderma reesei],3ZZ1_A Chain A, BETA-D-GLUCOSIDE GLUCOHYDROLASE [Trichoderma reesei] |
|
| 4.09e-92 | 31 | 687 | 11 | 713 | Chain A, Beta-D-glucoside glucohydrolase [Trichoderma reesei],4I8D_B Chain B, Beta-D-glucoside glucohydrolase [Trichoderma reesei] |
|
| 1.40e-91 | 33 | 578 | 23 | 640 | Chain A, Beta-glucosidase [Thermochaetoides thermophila] |
|
| 1.47e-88 | 28 | 681 | 4 | 706 | Crystasl Structure of Beta-glucosidase D2-BGL from Chaetomella Raphigera [Chaetomella raphigera] |
|
| 1.54e-88 | 33 | 583 | 43 | 664 | Chain A, Beta-glucosidase [Rasamsonia emersonii],5JU6_B Chain B, Beta-glucosidase [Rasamsonia emersonii],5JU6_C Chain C, Beta-glucosidase [Rasamsonia emersonii],5JU6_D Chain D, Beta-glucosidase [Rasamsonia emersonii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.63e-228 | 26 | 689 | 33 | 752 | Probable beta-glucosidase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=bglD PE=3 SV=2 |
|
| 2.64e-222 | 1 | 684 | 1 | 745 | Probable beta-glucosidase D OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglD PE=3 SV=2 |
|
| 3.31e-222 | 1 | 690 | 1 | 811 | Probable beta-glucosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglD PE=3 SV=2 |
|
| 1.06e-221 | 1 | 684 | 1 | 745 | Probable beta-glucosidase D OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglD PE=3 SV=1 |
|
| 1.11e-96 | 20 | 679 | 40 | 804 | Probable beta-glucosidase G OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
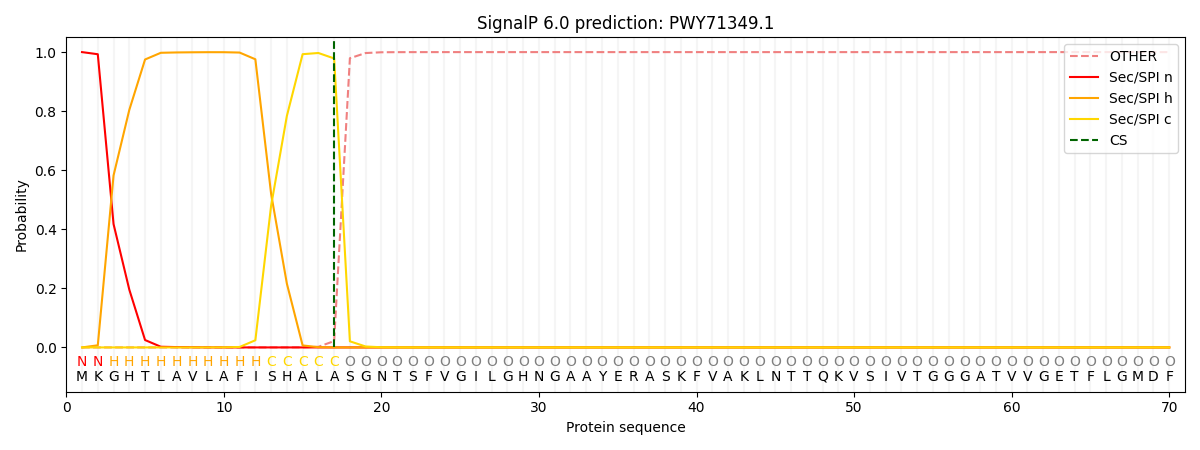
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000449 | 0.999526 | CS pos: 17-18. Pr: 0.9785 |
