You are browsing environment: FUNGIDB
CAZyme Information: PWY69453.1
You are here: Home > Sequence: PWY69453.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus eucalypticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus eucalypticola | |||||||||||
| CAZyme ID | PWY69453.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | CobW domain protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.67:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 77 | 424 | 9.1e-71 | 0.9569230769230769 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223597 | YejR | 2.73e-66 | 432 | 907 | 11 | 322 | GTPase, G3E family [General function prediction only]. |
| 349766 | CobW-like | 2.62e-58 | 432 | 676 | 10 | 198 | cobalamin synthesis protein CobW. The function of this protein family is unknown. The amino acid sequence of YjiA protein in E. coli contains several conserved motifs that characterizes it as a P-loop GTPase. YijA gene is among the genes significantly induced in response to DNA-damage caused by mitomycin. YijA gene is a homologue of the CobW gene which encodes the cobalamin synthesis protein/P47K. |
| 395231 | Glyco_hydro_28 | 8.94e-48 | 94 | 418 | 14 | 312 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| 396860 | cobW | 4.25e-47 | 432 | 661 | 10 | 179 | CobW/HypB/UreG, nucleotide-binding domain. This domain is found in HypB, a hydrogenase expression / formation protein, and UreG a urease accessory protein. Both these proteins contain a P-loop nucleotide binding motif. HypB has GTPase activity and is a guanine nucleotide binding protein. It is not known whether UreG binds GTP or some other nucleotide. Both enzymes are involved in nickel binding. HypB can store nickel and is required for nickel dependent hydrogenase expression. UreG is required for functional incorporation of the urease nickel metallocenter. GTP hydrolysis may required by these proteins for nickel incorporation into other nickel proteins. This family of domains also contains P47K, a Pseudomonas chlororaphis protein needed for nitrile hydratase expression, and the cobW gene product, which may be involved in cobalamin biosynthesis in Pseudomonas denitrificans. |
| 215540 | PLN03010 | 1.15e-30 | 104 | 425 | 106 | 391 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.08e-316 | 1 | 433 | 1 | 433 | |
| 2.08e-316 | 1 | 433 | 1 | 433 | |
| 2.21e-303 | 1 | 436 | 1 | 435 | |
| 2.21e-303 | 1 | 436 | 1 | 435 | |
| 1.42e-239 | 7 | 431 | 7 | 430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-49 | 61 | 436 | 27 | 387 | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis [Aspergillus tubingensis] |
|
| 6.55e-23 | 73 | 410 | 10 | 332 | Endopolygalacturonase from the phytopathogenic fungus Fusarium moniliforme [Fusarium verticillioides] |
|
| 2.69e-22 | 142 | 427 | 75 | 342 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
|
| 1.19e-21 | 142 | 427 | 75 | 342 | Chain A, endo-polygalacturonase [Evansstolkia leycettana] |
|
| 1.42e-21 | 142 | 427 | 67 | 334 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.92e-304 | 1 | 436 | 1 | 435 | Exopolygalacturonase B OS=Aspergillus niger OX=5061 GN=pgxB PE=3 SV=1 |
|
| 3.92e-304 | 1 | 436 | 1 | 435 | Probable exopolygalacturonase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pgxB PE=3 SV=1 |
|
| 2.53e-240 | 7 | 431 | 7 | 430 | Probable exopolygalacturonase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pgxB PE=3 SV=1 |
|
| 3.58e-240 | 7 | 431 | 7 | 430 | Probable exopolygalacturonase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pgxB PE=3 SV=2 |
|
| 1.43e-231 | 9 | 436 | 3 | 435 | Probable exopolygalacturonase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pgxB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
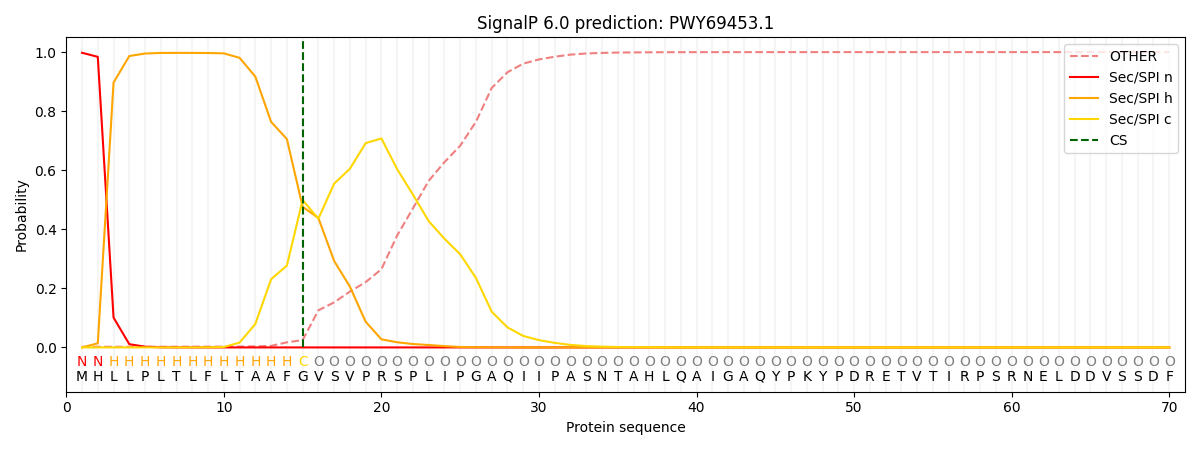
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.003741 | 0.996194 | CS pos: 15-16. Pr: 0.4989 |
