You are browsing environment: FUNGIDB
CAZyme Information: PWY69099.1
Basic Information
help
| Species |
Aspergillus heteromorphus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus heteromorphus
|
| CAZyme ID |
PWY69099.1
|
| CAZy Family |
AA7 |
| CAZyme Description |
cellulase family protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 412 |
MSFL01000034|CGC1 |
45743.97 |
4.0868 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AheteromorphusCBS117.55 |
11436 |
1448321 |
306 |
11130
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
33 |
367 |
4.4e-167 |
0.9970326409495549 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 395098
|
Cellulase |
1.62e-05 |
34 |
364 |
1 |
269 |
Cellulase (glycosyl hydrolase family 5). |
| 225344
|
BglC |
7.19e-04 |
25 |
403 |
29 |
394 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.03e-266 |
1 |
412 |
1 |
412 |
| 1.03e-266 |
1 |
412 |
1 |
412 |
| 1.74e-266 |
1 |
412 |
1 |
412 |
| 1.16e-212 |
15 |
412 |
15 |
412 |
| 5.73e-208 |
15 |
412 |
15 |
430 |
PWY69099.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.23e-52 |
3 |
399 |
13 |
388 |
Glycosyl hydrolase 5 family protein OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
| 2.85e-06 |
28 |
257 |
41 |
238 |
Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
This protein is predicted as SP
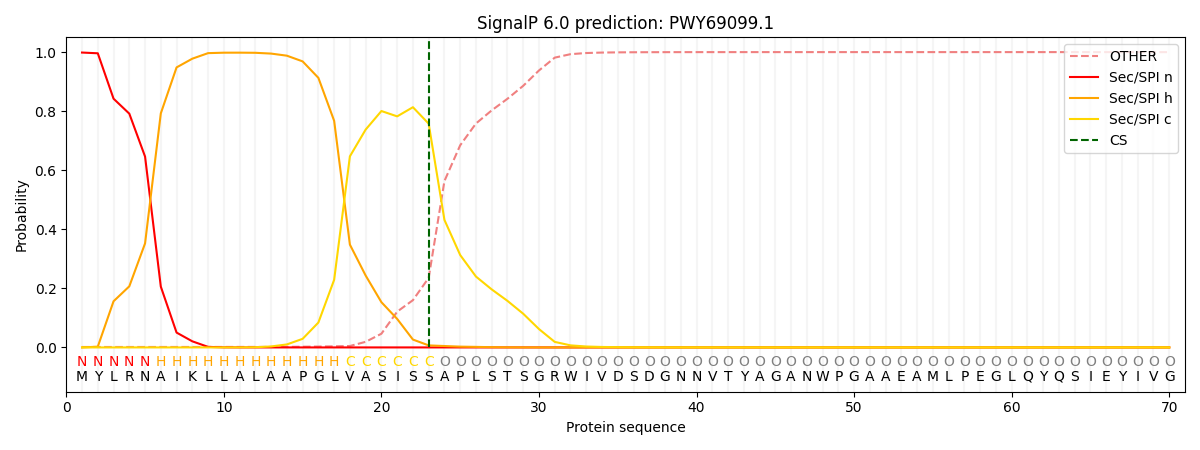
| Other |
SP_Sec_SPI |
CS Position |
| 0.002886 |
0.997103 |
CS pos: 23-24. Pr: 0.7576 |
There is no transmembrane helices in PWY69099.1.

